Megachile rotundata: Difference between revisions
Jump to navigation
Jump to search
| (102 intermediate revisions by the same user not shown) | |||
| Line 3: | Line 3: | ||
== Original Traces == | == Original Traces == | ||
* | * Illumina quality scores | ||
<pre style="background:yellow"> | <pre style="background:yellow"> | ||
lib insert mates reads readLen ~coverage(500M genome) adaptor repeat outies | |||
s_2_3kbp 3000 21,563,283 43,126,566 124 11 21%,19% 31% yes | |||
s_2_8kbp 8000 198,377 396,754 124 0.1 28%,29% 58% yes | |||
s_2_5kbp 5000 36,218,589 72,437,178 35 5 no ? yes | |||
s_3 475 35,548,153 71,096,306 124 18 no 50% | |||
s_4 475 35,471,044 70,942,088 124 18 | |||
s_5 475 35,616,846 71,233,692 124 18 | |||
s_6 475 35,303,840 70,607,680 124 18 | |||
s_7 475 34,893,313 69,786,626 124 18 | |||
subTotal . 234,813,445 469,626,890 . 98* | |||
s_2_1.1kb 1100 32,634,858 65,269,716 100 13 no 53% | |||
s_2_1.4kb 1500 50,861,645 101,723,290 100 20.3 no 54% | |||
s_7_8kb 8-10kb 25,328,718 50,657,436 125,100 11.4 21%,13% 39% yes | |||
s_7_5kb 5.3kb 29,111,787 58,223,574 36 4.2 no 23% yes GATCGGAAGAGC | |||
</pre> | </pre> | ||
* Location | * Location | ||
/fs/szattic-asmg5/Bees/Megachile_rotundata/ | /fs/szattic-asmg5/Bees/Megachile_rotundata/ | ||
/fs/szattic-asmg5/Bees/Megachile_rotundata/newLibrary/ | /fs/szattic-asmg5/Bees/Megachile_rotundata/newLibrary/ | ||
/fs/szattic-asmg5/Bees/Megachile_rotundata/newLibrary2/ | |||
* Ftp | * Ftp | ||
| Line 31: | Line 35: | ||
== Corrected/Addaptor-free Traces == | == Corrected/Addaptor-free Traces == | ||
* Sample 10K mates from each lib; compute quality, base composition stats | |||
Location: | |||
/nfshomes/dpuiu/Megachile_rotundata/original_sample | |||
== Corrected/Addaptor-free Traces == | |||
=== 1st run (2010 Summer) === | |||
* Mated ones | * Mated ones | ||
| Line 42: | Line 54: | ||
s_6 475 33,223,371 66,446,742 36,102,470 | s_6 475 33,223,371 66,446,742 36,102,470 | ||
s_7 475 32,647,890 65,295,780 36,102,370 | s_7 475 32,647,890 65,295,780 36,102,370 | ||
subTotal . 170,218,743 340,437,486 | |||
s_2_1.1kbp 1100 21,502,608 43,005,216 33,900,260 | |||
s_2_1.4kbp 14000 29,125,202 58,250,404 47,076,236 | |||
s_7_8kb 8-10kb ? ? | |||
* Subset | |||
lib mates reads repeatReads | lib mates reads repeatReads | ||
s_2_3kb.trim64 2,888,124(13.39% orig) 5,776,248 ~0 # these reads aligned to the all read SOAPdenovo assembly; ~ 20% of the mates have 1 mate aligned to linker & ~ 80% of mates are linker free | s_2_3kb.trim64 2,888,124(13.39% orig) 5,776,248 ~0 # these reads aligned to the all read SOAPdenovo assembly; ~ 20% of the mates have 1 mate aligned to linker & ~ 80% of mates are linker free | ||
| Line 60: | Line 75: | ||
/fs/szattic-asmg5/Bees/Megachile_rotundata/error_free/s_?_?_sequence.cor.txt # short insert libs | /fs/szattic-asmg5/Bees/Megachile_rotundata/error_free/s_?_?_sequence.cor.txt # short insert libs | ||
/fs/szattic-asmg5/Bees/Megachile_rotundata/frg/ # frg files to assemble | /fs/szattic-asmg5/Bees/Megachile_rotundata/frg/ # frg files to assemble | ||
=== 2nd run (2011 March) === | |||
* Mated ones | |||
lib insert mates mapped redundant | |||
s_2_8kb 8000 0 | |||
s_2_5kb 5000 0 | |||
s_3 450 33,044,498 | |||
s_4 450 33,243,183 | |||
s_5 450 33,164,796 | |||
s_6 450 33,227,529 | |||
s_7 32,652,698 | |||
s_2_1.1kb 1100 27,353,742 25.31% 2% | |||
s_2_1.4kb 1400 43,300,854 20% 3.7% | |||
s_2_3kb 3000 11,074,463 48.5% 27.3% | |||
s_7_5kb 5000 18,954,679 43% 27% | |||
s_7_8kb 8000 15,101,958 36.6% 35.9% | |||
* Location | |||
/fs/szattic-asmg5/Bees/Megachile_rotundata/error_correction_3-10/ # re-corrected traces (March 2011) | |||
ftp://ftp.cbcb.umd.edu/pub/data/assembly/Megachile_rotundata/reads.2011-03/ | |||
== Adaptors == | == Adaptors == | ||
!!! 5' & 3' linkers different than the Bumblebee ones. | |||
>circularizarion | >circularizarion | ||
CGTAATAACTTCGTATAGCATACATTATACGAAGTTATACGA | CGTAATAACTTCGTATAGCATACATTATACGAAGTTATACGA | ||
>circularizarion.revcomp | >circularizarion.revcomp | ||
TCGTATAACTTCGTATAATGTATGCTATACGAAGTTATTACG | TCGTATAACTTCGTATAATGTATGCTATACGAAGTTATTACG | ||
>5 | |||
CGATAACTTCGTATAATGTATGCTATACGAAGTTATTA | |||
>3 | |||
GCATAACTTCGTATAGCATACATTATACGAAGTTATACGA | |||
== Frequent kmers == | == Frequent kmers == | ||
| Line 94: | Line 139: | ||
ginkgo:/scratch1/dpuiu/Megachile_rotundata/Data/error_free/noRepeats/ # repeat free FASTQ reads & FRG files | ginkgo:/scratch1/dpuiu/Megachile_rotundata/Data/error_free/noRepeats/ # repeat free FASTQ reads & FRG files | ||
/fs/szattic-asmg5/Bees/Megachile_rotundata/error_free_repeats/ # repeat ids & unique FASTQ reads | /fs/szattic-asmg5/Bees/Megachile_rotundata/error_free_repeats/ # repeat ids & unique FASTQ reads | ||
== Linker removal == | |||
/fs/szdevel/dpuiu/removeLinkerFromMates/ | |||
== Identify duplicates == | |||
paste s_2_?_8kb*.txt | perl -ane 'chomp; if($.%4==1) { s/\@(\S+)[12]//; printf("%-45s",$1); } elsif($.%4==2) { print substr($F[0],0,32),"\t",substr($F[1],0,32),"\n" } ' | sort -k2,3 | ./getDuplicates.pl > s_2_8k.tab | |||
cat s_2_8k.tab | perl -ane 'print $F[0],"1\n";' > s_2_1_8k.redundant | |||
cat s_2_8k.tab | perl -ane 'print $F[0],"2\n";' > s_2_2_8k.redundant | |||
== Pacbio data == | |||
* [[Trace_formatting#Megachile_rotundata_PacBio_sequence]] | |||
* Ftp location: | |||
https://genomexfer.wustl.edu/gxfer1/3535746101561/ | |||
beiyohmaifie | |||
cuyogavomaij | |||
* Location | |||
/fs/szattic-asmg5/Bees/Megachile_rotundata/PacBio | |||
/fs/szattic-asmg5/Bees/Megachile_rotundata/PacBio/PRIMARY_DATA/*/*fasta # 95 files | |||
/fs/szattic-asmg5/Bees/Megachile_rotundata/PacBio/HQ_READS/*fasta # 85 files | |||
/fs/szattic-asmg5/Bees/Megachile_rotundata/PacBio/FILTERED_TRIMMED_READS/filtered_subreads.fa # 1 file | |||
* FASTA read length stats | |||
. elem min q1 q2 q3 max mean n50 sum | |||
PRIMARY_DATA 7138674 51 337 550 1054 15128 860 1268 6142469410 | |||
HQ_READS 858432 1 470 799 1291 10612 997 1336 855439838 | |||
FILTERED_TRIMMED_READS 1175261 1 168 517 863 6617 613 934 720454953 | |||
* FASTA read gc% stats | |||
. elem min q1 q2 q3 max mean n50 sum | |||
PRIMARY_DATA 7138674 0.20 54.11 62.95 72.37 98.97 62.97 65.85 . | |||
HQ_READS 858432 0.00 42.18 49.50 60.48 100.00 52.04 53.35 . | |||
FILTERED_TRIMMED_READS 1175261 0.00 36.01 42.86 48.44 100.00 41.35 44.80 . | |||
=== 1000 FILTERED_TRIMMED_READS sampled read stats === | |||
all 1000 | |||
bwa -b5 -q2 -r1 -z10 522 | |||
nucmer -l10 -c20 719 # 161 reads have less than 30bp aligned | |||
nucmer -l10 -c30 439 | |||
nucmer -l15 -c30 433 | |||
=== FILTERED_TRIMMED_READS read stats === | |||
= Assemblies = | = Assemblies = | ||
| Line 353: | Line 444: | ||
deg 258950 64 121 133 176 13861 162.26 161 42018113 | deg 258950 64 121 133 176 13861 162.26 161 42018113 | ||
utg 656004 64 84 124 162 128674 499.73 5935 327823531 | utg 656004 64 84 124 162 128674 499.73 5935 327823531 | ||
scfSeqContigs 7507 6 3643 10925 40664 567232 33678.11 93144 252821608 # gaps closed by SOAPGapCloser | |||
</pre> | </pre> | ||
| Line 543: | Line 637: | ||
=== Location === | === Location === | ||
mulberry:/scratch2/dpuiu/Megachile_rotundata/Assembly/wgs-noOBT/ | mulberry:/scratch2/dpuiu/Megachile_rotundata/Assembly/wgs-noOBT/ (to be deleted) | ||
/fs/szattic-asmg5/Bees/Megachile_rotundata/Assembly/wgs-noOBT/ | /fs/szattic-asmg5/Bees/Megachile_rotundata/Assembly/wgs-noOBT/ ** | ||
* Ftp | * Ftp | ||
| Line 559: | Line 653: | ||
-m <min> Number of mate samples to recompute an insert size, default is 100 => increase to ? | -m <min> Number of mate samples to recompute an insert size, default is 100 => increase to ? | ||
== SOAPdenovo (Tanja) | == SOAPdenovo (Tanja) == | ||
cat *.ContigIndex | grep -v ^E | grep -v ^i | count.pl -i 1 | getSummary.pl -j 1 -t "contigs" | cat *.ContigIndex | grep -v ^E | grep -v ^i | count.pl -i 1 | getSummary.pl -j 1 -t "contigs" | ||
| Line 566: | Line 660: | ||
* Stats | * Stats | ||
. elem min q1 q2 q3 max mean n50 sum | . elem min q1 q2 q3 max mean n50 sum | ||
scaf 7863 102 903 3272 17692 2,338,728** 37825 240,706 297423517 # N50 for Bee was 1.17M | scaf 7863 102 903 3272 17692 2,338,728** 37825 240,706 297423517 # N50 for Bee was 1.17M | ||
contigs 9742349 31 32 33 37 114,832 60 44 585430821 | contigs 9742349 31 32 33 37 114,832 60 44 585430821 | ||
contigs(>100bp) 177327 100 131 261 1398 114,832 1333 3897** 236496823 # N50 for Bee was 7K | contigs(>100bp) 177327 100 131 261 1398 114,832 1333 3897** 236496823 # N50 for Bee was 7K | ||
* Location | * Location | ||
| Line 580: | Line 672: | ||
=== Stats === | === Stats === | ||
. elem min q1 q2 q3 max mean n50 sum | . elem min q1 q2 q3 max mean n50 sum | ||
scaff 25,119 351 1896 4444 10914 1,102,803 11041 26876 277,338,897 | scaff 25,119 351 1896 4444 10914 1,102,803 11041 26876 277,338,897 | ||
scaff | scaff(all) 51,551 100 121 489 4320 1,102,830 5516 25940 284,368,749 | ||
scaffContigs(all) 263,977 33 78 149 878 121,523 914 3273 241,406,149 # <= scafffasta2fasta.pl | scaffContigs(all) 263,977 33 78 149 878 121,523 914 3273 241,406,149 # <= scafffasta2fasta.pl | ||
| Line 634: | Line 724: | ||
. elem min q1 q2 q3 max mean n50 sum | . elem min q1 q2 q3 max mean n50 sum | ||
scaff 24,602 333 1724 4380 11049 1,103,462 11135 27887 273963709 | |||
contigs(all) | scaff(all) 40,883 100 146 1366 5960 1,103,447 6833 27088 279355387 | ||
contigs(100bp+) | |||
scaffContigs(all) 231,559 33 75 153 982 148,167 1039 3950 240752397 | |||
contigs(all) 2,515,516 31 33 36 52 148,198 131 1880 330512911 | |||
contigs(100bp+) 184,395 100 127 235 1316 148,198** 1263 3730 232932308 | |||
=== Location === | === Location === | ||
| Line 642: | Line 736: | ||
ginkgo: /scratch1/dpuiu/Megachile_rotundata/Assembly/SOAPdenovo-partial.3.noRepeats/ | ginkgo: /scratch1/dpuiu/Megachile_rotundata/Assembly/SOAPdenovo-partial.3.noRepeats/ | ||
== SOAPdenovo ; s_2_3kb & s_ 2_8kb soap-aligned & trim64; s_[34567] no repeat ** == | === Aligning long inserts to this assembly ==== | ||
* Should trim the linker (mostly at the beginning of the reads ) | |||
show-coords -H 1000_12.filter-1.delta | sort -k19 | p '$F[18]=~s/[12]$//; print $p,$_,"\n" if($P[18] eq $F[18] and $P[17] ne $F[17]); $p=$_; @P=@F;' | pretty | |||
... | |||
cat *-1000*filter*delta | show-coords.pl | sort -k19 | ... | |||
417 469 | 2 54 | 53 53 | 98.11 | 2148 54 | 2.47 98.15 | scaffold16793 HWI-EAS385_0062:2:1:10306:11665#CAGATC/1 [CONTAINS] | |||
10634 10719 | 39 124 | 86 86 | 96.51 | 10732 124 | 0.8 69.35 | scaffold21864 HWI-EAS385_0062:2:1:10306:11665#CAGATC/2 . | |||
1 38 | 40 3 | 38 38 | 100 | 42 124 | 90.48 30.65 | linker.rev HWI-EAS385_0062:2:1:10306:11665#CAGATC/2 . | |||
1 38 | 40 3 | 38 38 | 100 | 42 124 | 90.48 30.65 | linker.fwd HWI-EAS385_0062:2:1:10759:13923#CAGATC/1 . | |||
1 82 | 41 122 | 82 82 | 95.12 | 7015 124 | 1.17 66.13 | scaffold5186 HWI-EAS385_0062:2:1:10759:13923#CAGATC/1 . | |||
5293 5416 | 1 124 | 124 124 | 94.35 | 7427 124 | 1.67 100 | scaffold7361 HWI-EAS385_0062:2:1:10759:13923#CAGATC/2 [CONTAINS] | |||
== SOAPdenovo ; s_2_3kb & s_ 2_8kb soap-aligned & trim64; s_2_1.1k & s_2_1.4k ; s_[34567] no repeat ** == | |||
=== Stats === | === Stats === | ||
<pre style="background:yellow"> | |||
. elem min q1 q2 q3 max mean n50 sum | |||
contigs | scaf 6242 228 640 2061 10681 3,022,211 42964 456,467 268,183,797 | ||
contigs( | scafSeq 17702 100 119 178 830 2,999,853 15136 452,601 267,954,678 | ||
contigs 4138734 32 32 34 41 111,437 94 1286 389,303,351 | |||
contigs100+ 181683 100 131 252 1385 111,437 1307 3789 237,563,819 | |||
scafSeqContigs 124749 2 90 466 2237 123,623 1970 5,800 245,850,885 | |||
scafSeqContigsClosed 25288 2 130 382 5576 626,941 10218 61,363 258,400,185 # after running GapCloser | |||
</pre> | |||
* GC contig cvg bias | |||
[[Image:Megachile.contig10000.png]] | |||
=== Scaffold links === | |||
perl ~/bin/remapSOAPscafId.pl *.scaf -p 5 | p 'print $_ unless($F[-2]=~/DOWN/ and $F[-1]=~/UP/);' | more | |||
... | |||
>2 76 50030 | |||
2.2 206 58 #DOWN #UP 543.152:5:1248 | |||
2.4 611 1458 #DOWN #UP 543.152:26:1477 | |||
2.24 18951 911 #DOWN 2268.26:31:139 #UP | |||
2.25 20219 2382 #DOWN #UP 2268.26:28:212 | |||
2.40 34053 150 #DOWN 1363.46:10:1041 #UP | |||
2.42 34434 76 #DOWN 1363.46:6:804 #UP | |||
2.43 34500 70 #DOWN 1363.46:6:682 #UP | |||
2.60 41566 200 #DOWN 808.70:8:219 #UP | |||
2.62 42030 1405 #DOWN #UP 808.70:9:82 | |||
.. | |||
>543 153 191782 | |||
543.63 108072 1765 #DOWN 158.305:5:198 #UP | |||
543.66 110202 77 #DOWN #UP 158.305:6:185 | |||
543.98 163144 810 #DOWN 1285.8:25:206 #UP | |||
543.99 164653 393 #DOWN #UP 91.485:6:197 | |||
543.121 176938 49 #DOWN #UP 985.12:24:88 | |||
543.125 178897 102 #DOWN 234.161:5:275 #UP | |||
543.128 179479 63 #DOWN 220.220:11:214 1951.10:14:284 576.114:16:256 #UP 576.116:8:474 234.161:8:178 | |||
543.152 188359 3153 #DOWN 2.4:26:1477 2.2:5:1248 #UP | |||
... | |||
=== SOAPdenovo vs CA === | |||
Aligns contigs 200+bp with nucmer.amos | |||
set REFN=`grep -c ">" ref.seq` | |||
@ REFN/=20 | |||
nucmer.amos -D REF=ref -D QRY=qry -D REFN=$REFN ref-qry | |||
. elem min q1 q2 q3 max mean n50 sum | |||
ref(CA) 16507 200 3678 7660 17785 317387 15436 31240 254794314 | |||
qry(SOAPdenovo) 100877 200 499 1224 2328 111437 2248 4103 226807646 | |||
cat ref-qry.delta | grep "^>" | sed 's/>//' | awk '{print $1,$3}' | sort -u | getSummary.pl -i 1 | |||
. elem min q1 q2 q3 max mean n50 sum | |||
ref-hits 16195 200 3816 7848 18116 317387 15711 31309 254444709 | |||
qry-hits 95343 200 610 1307 2427 111437 2356 4171 224616404 | |||
. elem min q1 q2 q3 max mean n50 sum | |||
ref-miss 312 200 244 483 1575 13984 1121 2262 349605 | |||
qry-miss 5534 200 235 298 442 5658 396 421 2191242 | |||
=== Location === | === Location === | ||
mulberry: /scratch2/dpuiu/Megachile_rotundata/Assembly/SOAPdenovo.noRepeats.trim64/ | mulberry: /scratch2/dpuiu/Megachile_rotundata/Assembly/SOAPdenovo.noRepeats.trim64/ (to be deleted) | ||
/fs/szattic-asmg5/Bees/Megachile_rotundata/Assembly/SOAPdenovo.noRepeats.trim64/ ** | |||
/fs/ftp-cbcb/pub/data/assembly/Megachile_rotundata.SOAPdenovo.2/ => ftp://ftp.cbcb.umd.edu/pub/data/assembly/Megachile_rotundata.SOAPdenovo.2/ | |||
== SOAPdenovo ; s_2_3kb & s_ 2_8kb soap-aligned & trim64; s_2_1.1k,s_2_1.4k,s_[34567] no repeat & trim64 == | |||
* Trimming all reads to the first 64bp does not change the results much (actually getting a little worse) | |||
=== Stats === | |||
. elem min q1 q2 q3 max mean n50 sum | |||
scaf 7615 441 896 2238 11462 2394735 37960 326734 289063269 | |||
scafSeq 23906 100 118 170 914 2396948 12271 324184 293340377 | |||
contigs 1738246 32 33 37 61 122972 174 2037 302039034 | |||
contigs100+ 214540 100 135 264 1263 122972 1110 2941 238063650 | |||
== SOAPdenovo K=31 (new data) == | |||
* Location | |||
/scratch1/dpuiu/Megachile_rotundata/Assembly/SOAPdenovo/K31 | |||
ftp://ftp.cbcb.umd.edu/pub/data/assembly/Megachile_rotundata.SOAPdenovo.2011-03/ | |||
* Assembly stats: | |||
. elem min q1 q2 q3 max mean n50 sum | |||
scf 16115 100 111 134 264 4173260 17307 1288790 278898652 | |||
scfLen 16115 100 111 134 208 3855147 14496 950327 233605903 | |||
ctg 8829648 32 32 34 39 121554 62 3201 553630969 | |||
scf2 16115 100 111 134 260 4033560 16539 1067152 266528571 | |||
scf2Len 16115 100 111 134 259 4004254 16236 1071079 261641263 | |||
ctg2 26490 3 120 243 5253 520023 9877 64739 261641355 | |||
cat asm.K31.peGrads | tail -6 | p 'print $F[0], " ", $F[1]-$P[1],"\n"; @P=@F' | pretty | |||
350 330665408 | |||
1100 54707484 | |||
1400 86601708 | |||
3000 22148926 | |||
5300 37909358 | |||
8000 30203916 | |||
cat asm.K31.links | awk '{print $5}' | uniq -c | awk '{print $2,$1}' | |||
350 7375561 | |||
1100 579996 | |||
1400 604951 | |||
3000 340192 | |||
5300 669339 | |||
8000 184868 | |||
7375561 350 | |||
579996 1100 | |||
604951 1400 | |||
340192 3000 | |||
669339 5300 | |||
184868 8000 | |||
== SOAPdenovo K=47 (new data) ** == | |||
<pre style="background:yellow"> | |||
. elem min q1 q2 q3 max mean n50 sum | |||
scaf 3495 259 408 757 4763 6,173,378 82382 2,124,089 287,925,734 | |||
scafSeq 31774 100 110 128 172 6,174,792 9215 2,124,853* 292,784,153 | |||
scafSeq2 31774 100 110 128 172 5,876,085 8689 1,814,396 276,082,351 | |||
contigs 10217806 48 48 50 56 175,671 77 4,701 792,925,336 | |||
contigs100+ 224315 100 126 177 933 175,671 1134 4,701 254,309,160 | |||
scafSeqContigsClosed 43232 2 114 147 740 479,105 6228 63,194* 269,243,097 | |||
</pre> | |||
Location: | |||
/fs/szattic-asmg5/Bees/Megachile_rotundata/Assembly/SOAPdenovo.10libs.K47 | |||
=== SOAPdenovo vs CA === | |||
elem min q1 q2 q3 max mean n50 sum | |||
CA 16848 64 3501 7433 17428 317387 15124 31240 254808119 | |||
SO 43232 2 114 147 740 479105 6228 63194 269243097 | |||
CA.no_hits 315 64 124 150 188 1353 168 167 52986 | |||
SO.no_hits 29679 2 109 123 151 5766 185 158 5489188 | |||
= Allpaths-LG (experiment) = | |||
* Shred SOAPdenovo K=47 contigs >=180bp ; use them as fragment library | |||
. elem min q1 q2 q3 max mean n50 sum | |||
contig.180+ 110580 180 264 964 2321 175671 2166 4701 239535581 | |||
* Libraries used: (in_libs.csv) | |||
library_name, project_name, organism_name, type, paired, frag_size, frag_stddev, insert_size, insert_stddev, read_orientation, genomic_start, genomic_end #mates | |||
frag, genome, genome, fragment, 1, 180, 20, , , inward, 0, 0 22,024,446 # originally 176,833,196 fragments insert_size=475bp | |||
s_2_1.1kb, genome, genome, jumping, 1, , , 1100, 110, inward, 0, 0 32,634,858 | |||
s_2_1.4kb, genome, genome, jumping, 1, , , 1400, 140, inward, 0, 0 50,861,645 | |||
s_7_5kb, genome, genome, jumping, 1, , , 5300, 530, outward, 0, 0 29,111,787 | |||
s_7_8kb, genome, genome, jumping, 1, , , 8000, 800, outward, 1, 37 25,328,718 | |||
type #mates.original #mates.corrected | |||
frag 22,024,446 21,925,564 | |||
jumping 137,937,008 5,465,197 | |||
* Assembly stats: | |||
<pre style="background:yellow"> | |||
. elem min q1 q2 q3 max mean n50 sum | |||
scf 1964 1 1187 1550 6679 6,117,056 128,398 1,806,225 252,173,219 | |||
ctg 36959 1 1836 3140 6343 278,684 6,061 8,784 224,007,758 | |||
</pre> | |||
= Genbank submission = | |||
* [http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?id=143995 Taxonomy] | |||
* [http://www.ncbi.nlm.nih.gov/genomes/mpfsubmission.cgi Project registration] | |||
* [http://www.ncbi.nlm.nih.gov/genomes/mpfsubmission.cgi?show=952ECB8E-7946-4B90-B982-4C2B350C171F Project ID 66515] | |||
* Information | |||
Project Type: Single Species Project | |||
Contacts: Daniela Puiu dpuiu@umiacs.umd.edu, Steven Salzberg salzberg@umiacs.umd.edu | |||
Submitting Organization: University of Illinois & University of Maryland | |||
Sequencing Center: Keck Center for Comparative and Functional Genomics, University of Illinois BCUI Biotechnology Center, Univ. Illinois (BCUI) | |||
Consortium Name: Alfalfa Leafcutter Bee Genome Consortium | |||
Organism Name: Megachile rotundata | |||
Strain/isolate/breed: North American commercial strain | |||
Locus Tag Prefix: MROT #3+ letters | |||
Source of DNA used for sequencing: whole body, haploid brother males | |||
Sequencing Method: wgs | |||
Sequencing Technology: Illumina | |||
Estimated Genome Size: 250Mb # the haploid genome size | |||
Brief description of the importance: Megachile rotundata, alfalfa leafcutting bee, is a solitary bee species. It is the #3 agricultural pollinator in the United States and is commercially managed for alfalfa seed production. | |||
Comments to the staff: | |||
DNA | |||
whole genome sequencing | |||
single genome; | |||
no annotation | |||
assembly name MROT_1.0 | |||
assembly method: SOAPdenovo Assembler v_1.05 | |||
plan to update: ? | |||
expect to release : ? | |||
strain information to be submitted soon | |||
genome coverage: 300x | |||
sequencing technology: Illumina GA IIx ?? | |||
Author list: | |||
Gene E. Robinson, | |||
Hugh M. Robertson, | |||
Matthew E. Hudson, | |||
Kim Walden, | |||
Brielle J. Fischman, | |||
Theresa Pitts-Singer, | |||
Rosalind James | |||
Steven Salzberg, | |||
Daniela Puiu, | |||
Tanja Magoc, | |||
David Kelley, | |||
Aleksey Zimin , | |||
* Best assembly : SOAPdenovo K=47 | |||
/fs/szattic-asmg5/Bees/Megachile_rotundata/Assembly/SOAPdenovo.10libs.K47/genbank[1-3] | |||
* Sequence length statistics [1]: | |||
number min max mean n50 sum | |||
scaffolds 31774 100 5876085 8689 1814396 276082351 | |||
contigs 42781 100 479105 6293 69010 269224383 | |||
* Removed 6 contaminants & scaff < 200bp [2] | |||
. elem min max mean n50 sum | |||
scaffolds 6367 200 5876085 42857 1699680 272873468 | |||
contigs 17374 100 479105 15311 64153 266015500 | |||
* NCBI comments: '' There are 668 contigs that are <200bp remaining. Some of these are internal components of scaffolds, but many of these are at the ends of scaffolds so should be removed. For example, 249 of them are the first or component of a scaffold or the only component of a singleton scaffold (list.ShortTerminalContigs). And some of them are the only components of a multi-component scaffold, eg these two scaffolds are made entirely of short contigs ... '' | |||
. elem min max mean n50 sum | |||
scaffolds 6266 200 5876085 43514 1814396 272660569 # 101 scaffolds deleted | |||
contigs 16706 200 479105 15917 69010 265916502 # 668 contigs deleted | |||
* GPID Organism name Accession | |||
----------------------------------------------- | |||
66515 Megachile rotundata AFJA00000000 | |||
Please cite the accession number as usual: | |||
This Whole Genome Shotgun project has been deposited at | |||
DDBJ/EMBL/GenBank under the accession AFJA00000000. | |||
The version described in this paper is the first version, | |||
AFJA01000000. | |||
Latest revision as of 20:22, 21 July 2011
Data
Original Traces
- Illumina quality scores
lib insert mates reads readLen ~coverage(500M genome) adaptor repeat outies
s_2_3kbp 3000 21,563,283 43,126,566 124 11 21%,19% 31% yes
s_2_8kbp 8000 198,377 396,754 124 0.1 28%,29% 58% yes
s_2_5kbp 5000 36,218,589 72,437,178 35 5 no ? yes
s_3 475 35,548,153 71,096,306 124 18 no 50%
s_4 475 35,471,044 70,942,088 124 18
s_5 475 35,616,846 71,233,692 124 18
s_6 475 35,303,840 70,607,680 124 18
s_7 475 34,893,313 69,786,626 124 18
subTotal . 234,813,445 469,626,890 . 98*
s_2_1.1kb 1100 32,634,858 65,269,716 100 13 no 53%
s_2_1.4kb 1500 50,861,645 101,723,290 100 20.3 no 54%
s_7_8kb 8-10kb 25,328,718 50,657,436 125,100 11.4 21%,13% 39% yes
s_7_5kb 5.3kb 29,111,787 58,223,574 36 4.2 no 23% yes GATCGGAAGAGC
- Location
/fs/szattic-asmg5/Bees/Megachile_rotundata/ /fs/szattic-asmg5/Bees/Megachile_rotundata/newLibrary/ /fs/szattic-asmg5/Bees/Megachile_rotundata/newLibrary2/
- Ftp
ftp.biotec.illinois.edu ftp://username@ftp.biotec.illinois.edu login: generobi Password: GRbeehi3
Corrected/Addaptor-free Traces
- Sample 10K mates from each lib; compute quality, base composition stats
Location: /nfshomes/dpuiu/Megachile_rotundata/original_sample
Corrected/Addaptor-free Traces
1st run (2010 Summer)
- Mated ones
lib insert mates reads repeatReads s_2_3kb 3000 4,823,235 (22%orig) 9,646,470 4,349,208 (45%) s_2_8kb 8000 111,267 (56%orig) 222,534 167,246 (75%) s_2_5kb 5000 36,218,589 72,437,178 35 # same as original s_3 475 33,024,597(92%orig) 66,049,194 35,777,342 (54%) s_4 475 33,237,593 66,475,186 36,247,656 s_5 475 33,150,790 66,301,580 36,350,706 s_6 475 33,223,371 66,446,742 36,102,470 s_7 475 32,647,890 65,295,780 36,102,370 subTotal . 170,218,743 340,437,486 s_2_1.1kbp 1100 21,502,608 43,005,216 33,900,260 s_2_1.4kbp 14000 29,125,202 58,250,404 47,076,236 s_7_8kb 8-10kb ? ?
- Subset
lib mates reads repeatReads s_2_3kb.trim64 2,888,124(13.39% orig) 5,776,248 ~0 # these reads aligned to the all read SOAPdenovo assembly; ~ 20% of the mates have 1 mate aligned to linker & ~ 80% of mates are linker free s_2_8kb.trim64 4,883(0.01% orig) 9,766 ~0 # these reads aligned to the all read SOAPdenovo assembly
- repeatReads:
- at least one of the mate contains a perfect match of one of the 15 frequent 22mers listed below
- 32.5%GC in repeatREads vs ~ 35.5%GC in uniqueReads
- Location
/fs/szattic-asmg5/Bees/Megachile_rotundata/error_correction/large_libs/s_?_?_?kb.sequence.cor.all.txt # large insert libs ; inverted compared to the original (outies => innies) /fs/szattic-asmg5/Bees/Megachile_rotundata/error_free/s_?_?_sequence.cor.txt # short insert libs /fs/szattic-asmg5/Bees/Megachile_rotundata/frg/ # frg files to assemble
2nd run (2011 March)
- Mated ones
lib insert mates mapped redundant s_2_8kb 8000 0 s_2_5kb 5000 0 s_3 450 33,044,498 s_4 450 33,243,183 s_5 450 33,164,796 s_6 450 33,227,529 s_7 32,652,698 s_2_1.1kb 1100 27,353,742 25.31% 2% s_2_1.4kb 1400 43,300,854 20% 3.7%
s_2_3kb 3000 11,074,463 48.5% 27.3% s_7_5kb 5000 18,954,679 43% 27% s_7_8kb 8000 15,101,958 36.6% 35.9%
- Location
/fs/szattic-asmg5/Bees/Megachile_rotundata/error_correction_3-10/ # re-corrected traces (March 2011) ftp://ftp.cbcb.umd.edu/pub/data/assembly/Megachile_rotundata/reads.2011-03/
Adaptors
!!! 5' & 3' linkers different than the Bumblebee ones.
>circularizarion CGTAATAACTTCGTATAGCATACATTATACGAAGTTATACGA >circularizarion.revcomp TCGTATAACTTCGTATAATGTATGCTATACGAAGTTATTACG >5 CGATAACTTCGTATAATGTATGCTATACGAAGTTATTA >3 GCATAACTTCGTATAGCATACATTATACGAAGTTATACGA
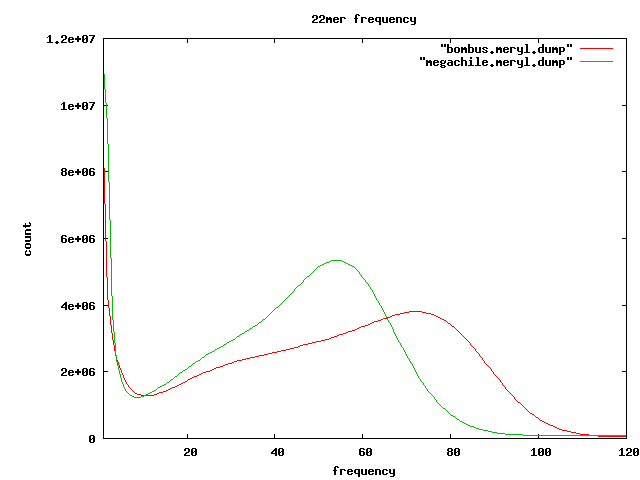
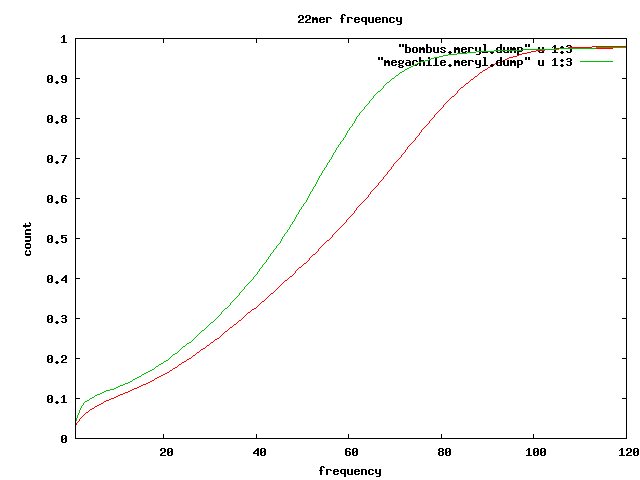
Frequent kmers
- 22mers which seem to appear in tandem
~ %reads
s_2_3kb s_2_8kb s_[4567] s_2_1k
-------------------------
1 AATCATACAATCACAATCATAC|GTATGATTGTGATTGTATGATT 12.04 20.1 9.99 21.18
2 CAATCACAATCATACAATCACA|TGTGATTGTATGATTGTGATTG 10.5 17.87 8.3 2.33
3 AATAATATGAGTTAGATTGATA|TATCAATCTAACTCATATTATT 7.94 11.77 21.47 9.12
4 AGTAATTGTCGTTCTATCGATC|GATCGATAGAACGACAATTACT 5.08 7.47 13.04 6.33
5 ATATAAGCATAATATGGCTAAT|ATTAGCCATATTATGCTTATAT 5.01 7.55 15.15 4.19
6 CACACAATCACACAATCACACA|TGTGTGATTGTGTGATTGTGTG 4.72 8.57 2.32
7 ATTACTCTTATTATTATCAATC|GATTGATAATAATAAGAGTAAT 4.62 6.67 11.8
8 TCACACAATCACAATCACACAA|TTGTGTGATTGTGATTGTGTGA 3.76 7.01 1.54
9 ACAATTACTATACTTATTACTC|GAGTAATAAGTATAGTAATTGT 2.94 4.39 8.46
10 AGACAGAGACAGAGACAGAGAC|GTCTCTGTCTCTGTCTCTGTCT 2.17 5.66 1.03 1.03
11 CACAATCACGATCACACAATCA|TGATTGTGTGATCGTGATTGTG 1.43 2.25 0.5
12 CTGTCTCTGTCTGTCTCTGTCT|AGACAGAGACAGACAGAGACAG 1.34 3.77 0.68
13 CAGCGGATATGTGCGAATTAGA|TCTAATTCGCACATATCCGCTG 0.8 0.54 0.73
14 CTGAGCACAATTCAACACCACA|TGTGGTGTTGAATTGTGCTCAG 0.58 0.35 0.68
15 AACCTAACCTAACCTAACCTAA|TTAGGTTAGGTTAGGTTAGGTT 0.06 0.15 0.03
total 31 55 50 52
- Location
ginkgo:/scratch1/dpuiu/Megachile_rotundata/Data/error_free/noRepeats/ # repeat free FASTQ reads & FRG files /fs/szattic-asmg5/Bees/Megachile_rotundata/error_free_repeats/ # repeat ids & unique FASTQ reads
Linker removal
/fs/szdevel/dpuiu/removeLinkerFromMates/
Identify duplicates
paste s_2_?_8kb*.txt | perl -ane 'chomp; if($.%4==1) { s/\@(\S+)[12]//; printf("%-45s",$1); } elsif($.%4==2) { print substr($F[0],0,32),"\t",substr($F[1],0,32),"\n" } ' | sort -k2,3 | ./getDuplicates.pl > s_2_8k.tab
cat s_2_8k.tab | perl -ane 'print $F[0],"1\n";' > s_2_1_8k.redundant
cat s_2_8k.tab | perl -ane 'print $F[0],"2\n";' > s_2_2_8k.redundant
Pacbio data
- Ftp location:
https://genomexfer.wustl.edu/gxfer1/3535746101561/ beiyohmaifie cuyogavomaij
- Location
/fs/szattic-asmg5/Bees/Megachile_rotundata/PacBio /fs/szattic-asmg5/Bees/Megachile_rotundata/PacBio/PRIMARY_DATA/*/*fasta # 95 files /fs/szattic-asmg5/Bees/Megachile_rotundata/PacBio/HQ_READS/*fasta # 85 files /fs/szattic-asmg5/Bees/Megachile_rotundata/PacBio/FILTERED_TRIMMED_READS/filtered_subreads.fa # 1 file
- FASTA read length stats
. elem min q1 q2 q3 max mean n50 sum PRIMARY_DATA 7138674 51 337 550 1054 15128 860 1268 6142469410 HQ_READS 858432 1 470 799 1291 10612 997 1336 855439838 FILTERED_TRIMMED_READS 1175261 1 168 517 863 6617 613 934 720454953
- FASTA read gc% stats
. elem min q1 q2 q3 max mean n50 sum PRIMARY_DATA 7138674 0.20 54.11 62.95 72.37 98.97 62.97 65.85 . HQ_READS 858432 0.00 42.18 49.50 60.48 100.00 52.04 53.35 . FILTERED_TRIMMED_READS 1175261 0.00 36.01 42.86 48.44 100.00 41.35 44.80 .
1000 FILTERED_TRIMMED_READS sampled read stats
all 1000 bwa -b5 -q2 -r1 -z10 522 nucmer -l10 -c20 719 # 161 reads have less than 30bp aligned nucmer -l10 -c30 439 nucmer -l15 -c30 433
FILTERED_TRIMMED_READS read stats
Assemblies
- CA Version: 6.1 (09/01/2010) /fs/szdevel/dpuiu/SourceForge/wgs-6.1/Linux-amd64/bin/runCA
- SOAP version 1.04: /nfshomes/dpuiu/szdevel/SOAPdenovo_Release1.04/
CA noOBT ; partial s_2_3kb, s_2_8kb, s_3
- Data : 3 libs : ~ 16X cvg
- Files
/fs/szattic-asmg5/Bees/Megachile_rotundata/Assembly/assemblyAdaptorFree/longLibrariesAdaptorFree/s_2_?kb_?.filter.fastq # inverted /fs/szattic-asmg5/Bees/Megachile_rotundata/error_free_better/s_3_?_sequence.cor.txt
Gatekeeper
LibraryName numActiveFRG numDeletedFRG numMatedFRG readLength clearLength GLOBAL 72,995,448 0 70632632 8307194830 8278360381 LegacyUnmatedReads 0 0 0 0 0 s_2_3kb 9,166,343 0 8736228 942501164 914798596 s_2_8kb 210,266 0 199620 21669112 20742291 s_3 63,618,839 0 61696784 7343024554 7342819494
UID IID mateUID mateIID libUID libIID isDel isNonRandom Orient Length clrBeginLATEST clrEndLATEST 110000000001 1 120000000001 2 s_2_3kb 1 0 0 I 75 0 75 120000000001 2 110000000001 1 s_2_3kb 1 0 0 I 123 0 123 110000000003 3 120000000003 4 s_2_3kb 1 0 0 I 90 0 90 120000000003 4 110000000003 3 s_2_3kb 1 0 0 I 123 40 123 ... 110009166343 9166343 0 0 s_2_3kb 1 0 0 U 76 11 76 210009166344 9166344 220009166344 9166345 s_2_8kb 2 0 0 I 123 21 123 ... 210009376609 9376609 0 0 s_2_8kb 2 0 0 U 88 0 88 320009376610 9376610 0 0 s_3 3 0 0 U 72 0 72 ... 310072995448 72995448 0 0 s_3 3 0 0 U 68 0 68
BOG/ tigStore
- Number of tigs in the store
tigStore -g asm.gkpStore -t asm.tigStore 2 -D unitiglist | tail -1 | awk '{print $1}' # 36318422
- Single read tigs
tigStore -g asm.gkpStore -t asm.tigStore 2 -U -d layout | grep -c '^data.num_frags 1$' # 34985292 ts2lay | grep -B 9 -A 3 '^data.num_frags 1$'
Stats
. elem min q1 q2 q3 max mean n50 sum #repeats comments scf 20,827 122 3228 6374 13700 202495* 11508 20462* 239696810 SOAPdenovo: max=1102803 , N50=26876 ctg 37,494 65 2185 3998 7706 191323* 6380 10151* 239226293 206 SOAPdenovo: max=121554 , N50=3138 deg 1,136,469 64 123 143 184 5031 160 164 181954480 807132 utg 1,437,146 64 123 143 195 67048 308 870 443759899 readsTotal 72,995,448 readsInContigs 27,837,956 readsInDegenerates 9,627,122 singletons 34,881,692 (47%) readsWithOuttieMate 3,028,956(4.15%) ???
Placed reads . badLong badOuttie badSame bothDegen bothSurrogate diffScaffold good notMated oneChaff oneDegen oneSurrogate s_2_3kb 534 2,998,286 458 1614846 9892 21872 27308 267328 979980 760044 65268 s_2_8kb 4 26,864 10 38636 114 294 178 5044 35465 7848 1022 s_3 11072 3,806 1104 2369982 61236 53370 23058022 1208112 3967689 371538 87260
Chaff reads . bothChaff notMated oneChaff s_2_3kb 1,277,760 162,787 979,980 s_2_8kb 53,588 5,602 35,465 s_3 27,684,878 713,943 3,967,689
Issues
- reads are renamed : HWI-EAS385_0062:2:1:1036:15608#GCCAAT/1 => UID:110000000001 => IID:1
- reads < 64bp are deleted from the beginning : ID mapping ???
- lib s_2 orientation ??? Too many badOuttie's
Location
ginkgo:/scratch1/dpuiu/Megachile_rotundata/Assembly/wgs-noOBT-partial.1/
CA noOBT ; partial : s_2_3kb, s_2_8kb, s_3 ; no repeats
- Reads that contain at least one of the 15 most frequent 22mers are deleted from the input set
Gatekeeper
LibraryName numActiveFRG numDeletedFRG numMatedFRG readLength clearLength GLOBAL 33811292 0 32385550 3781368484 3772837304 LegacyUnmatedReads 0 0 0 0 0 s_2_3kb 5103461 0 4928188 518415011 510187775 s_2_8kb 53215 0 51436 5395700 5289866 s_3 28654616 0 27405926 3257557773 3257359663
Overlapper
- Dirty 3' ends for the s_2_* reads
totalOvl avgOvl s_2_3kb 5' 4955294 9 s_2_3kb 3' 4955294 7 s_2_8kb 5' 51050 10 s_2_8kb 3' 51050 7 s_3 5' 27721948 9 s_3 3' 27721948 9
Bog
cat 4-unitigger/asm.cga.0 | head
Global Arrival Rate: 0.125220
There were 1,983,199 unitigs generated.
Unitig Length
65407 - 67872: 4
50209 - 58608: 5
40073 - 49263: 27
30132 - 39913: 72
20030 - 29892: 319
10001 - 19992: 1979
9007 - 9999: 673
8000 - 8999: 934
7000 - 7999: 1332
6000 - 6998: 2048
5000 - 5999: 3103
4000 - 4999: 4898
3000 - 3999: 8120
2000 - 2999: 14634
1000 - 1999: 26621
900 - 999: 4116
800 - 899: 4457
700 - 799: 5042
600 - 699: 6146
500 - 599: 8107
400 - 499: 11901
300 - 399: 19373
200 - 299: 64394
100 - 199: 1173219
90 - 99: 161987
80 - 89: 189874
70 - 79: 132943
64 - 69: 82098
UTG
- The default unitigger tried as well. Fails with the following message:
unitigger: AS_FGB_io.C:338: void add_overlap_to_graph(Aedge, Tfragment*, Tedge*, IntFragment_ID*, VarArrayIntEdge_ID*, int, int, int, IntEdge_ID*, IntEdge_ID*, IntEdge_ID*): Assertion `ialn > iahg' failed. Failure message: failed to unitig
Stats
- Larger max scf & ctg !!! (compared with "CA noOBT partial" that assembled the repeats as well)
. elem min q1 q2 q3 max mean n50 sum scf 21041 65 3174 6334 13482 337719* 11376 20153* 239366537 ctg 37668 65 2181 3963 7687 191376* 6343 10083* 238928665 deg 380596 64 107 126 170 4688 163 160 62151395 # ~22% of deg align with 1 mismatch to kmers utg 652051 64 115 133 225 67870 491 2469 320381694 readsTotal 33,811,292 readsInContigs 27,753,101 (82.08%) readsInDegenerates 4,004,853 (11.84%) singletons 1,276,811 (3.78%) # about 12% of singletons align with 1 mismatch to the frequent kmers
Placed reads . badLong badOuttie badSame bothDegen bothSurrogate diffScaffold good notMated oneChaff oneDegen oneSurrogate 1 582 2992742 410 773006 13486 20146 26870 159957 107838 753916 78412 2 1228 2 9486 84 124 62 1550 12940 7750 860 3 11354 3884 1074 2266364 90824 56066 23001316 1165421 346169 416116 156218
~/bin/asm2mdi.pl < asm.asm s_2_3kb 16 87 ??? s_2_8kb 8000 800 s_3 337 27
Location
ginkgo:/scratch1/dpuiu/Megachile_rotundata/Assembly/wgs-noOBT-partial.1.noRepeats/
CA noOBT ; partial : s_2_3kb & s_2_8kb (trim 64), s_3 no repeats
s_2_3kb & s_2_8kb reads filtering:
- trimmed to the first 64bp
- aligned to one of the SOAPdenovo assemblies (soap2)
- filtered only the mated reads and the single reads with mates aligned to different scaffolds.
The main goal was to get rid of the short inserts present in the long insert libraries (confuse the Celera scaffolder). At the same time I got rid of the linkers (since the linker did not get assembled by SOAPdenovo & all these reads align to the assembly) ...
LibraryName numActiveFRG numDeletedFRG numMatedFRG readLength clearLength GLOBAL 34220457 0 32793056 3613771597 3613573487 s_3 28654616 0 27405926 3257557773 3257359663 s_2_1_3kb.trim64 5556371 0 5377908 355607744 355607744 s_2_1_8kb.trim64 9470 0 9222 606080 606080
Stats
. elem min q1 q2 q3 max mean n50 sum scf 6,407 70 3609 10907 37417 854,387 38538 126946 246918298 ctg 46,442 64 882 2719 6385 184,930 5235.23 11002 243134341 deg 217,175 64 109 134 187 17,896 174.34 184 37861920 utg 564,404 64 77 124 226 82,947 539.31 3168 304389506 singletonReads 1,125,209
Location
ginkgo:/scratch1/dpuiu/Megachile_rotundata/Assembly/wgs-noOBT.partial.1.trim64/
CA noOBT ; partial : s_2_3kb, s_2_8kb, s_3 ; no repeats; reverse
- Reads that contain at least one of the 15 most frequent 22mers are deleted from the input set
- s_2_3kb & s_2_8kb libraries were reversed (since most of the reads in "CA noOBT partial" were outies)
- fewer bad mates
- Smaller contigs & scaffolds
CA noOBT ; partial : s_2_3kb, s_2_8kb, s_3 ; no repeats; no bad links
- Reads that contain at least one of the 15 most frequent 22mers are deleted from the input set
- s_2_3kb & s_2_8kb libraries : all mates listed as bad got "broken"
CA OBT ; partial : s_2_3kb, s_2_8kb, s_3 ; no repeats ; doDeduplication
- smaller contigs, scaffolds
CA noOBT ; partial s_3 , s_4 ; no repeats
- Reads that contain at least one of the 15 most frequent 22mers are deleted from the input set
Gatekeeper
LibraryName numActiveFRG numDeletedFRG numMatedFRG readLength clearLength GLOBAL 56839966 0 54032986 6425669702 6425397526 LegacyUnmatedReads 0 0 0 0 0 s_3 28654616 0 27405926 3257557773 3257359663 s_4 28185350 0 26627060 3168111929 3168037863
Stats
. elem min q1 q2 q3 max mean n50 sum scf 12908 148 4003 8811 22202 511831 19416 42207 250623581 ctg 23116 64 2888 5959 13088 255301 10828 20109 250302752 deg 274961 64 124 139 182 4652 172 164 47499725 utg 574873 64 123 132 202 128547 563 4478 324059992
. elem min q1 q2 q3 max mean n50 sum SLK 243 -50905 -7731 -448 -152 126 -5863 126 -1424832 CLK 516105 -245237 -59 36 82 208 -1112 208 -574243691 ULK 1123683 -150489 -83 6 71 209 -329 209 -369827832 CTP 10012 -19762 -20 -9 10 9876 -2 9876 -25270
Location
ginkgo:/scratch1/dpuiu/Megachile_rotundata/Assembly/wgs-noOBT-partial.2.noRepeats
CA noOBT ; partial : s_2_3kb & s_2_8kb (trim 64), s_3 ,s_4 no repeats
. elem min q1 q2 q3 max mean n50 sum scf 5159 64 3501 9986 45329 1493864 49143.80 180907 253532854 ctg 23598 64 1641 4533 12544 298755 10661.13 26042 251581405 deg 258950 64 121 133 176 13861 162.26 161 42018113 utg 656004 64 84 124 162 128674 499.73 5935 327823531 scfSeqContigs 7507 6 3643 10925 40664 567232 33678.11 93144 252821608 # gaps closed by SOAPGapCloser
Location:
ginkgo:/scratch1/dpuiu/Megachile_rotundata/Assembly/wgs-noOBT-partial.2.noRepeats.trim64/
CA noOBT **
- Data : 7 libs : ~ 74X cvg
Gatekeeper
LibraryName numActiveFRG numDelFRG numMatedFRG readLength clearLengt #repeats GLOBAL 326,236,387 0 315518526 37451489553 37418130441 s_2_3kb 9107424 0 9107424 942165284 910444046 # s_2_8kb 209336 0 209336 21814418 20787384 # s_3 63618839 0 61696784 7343024554 7342819494 # s_4 63544688 0 61255960 7291557748 7291478152 # s_5 63370860 0 61084368 7271218123 7271051639 # s_6 63780887 0 61685156 7359094156 7359012512 # s_7 62604353 0 60479498 7222615270 7222537214 #
Meryl
meryl -Dh -s 0-mercounts/asm-C-ms22-cm0 Found 30570218845 mers. Found 271464470 distinct mers. Found 11164787 unique mers. Largest mercount is 87984949; 1896 mers are too big for histogram. 1 11164787 0.0411 0.0004 2 9376915 0.0757 0.0010 3 3714582 0.0894 0.0013 ... 54 5344148 0.6573 0.1788 ... 87984949 1 # AATCATACAATCACAATCATAC
Overlap
- job count :
cat 1-overlapper/ovlopts.pl | grep ^\"h | wc -l 924
- Failures: 709 jobs failed; runCA 6.1 could not restart overlap properly !!!
cat 1-overlap/overlap*out | grep "^Could not" | sort -u Could not malloc memory (1305184948 bytes)
- Only ~ 60% of the reads had overlaps
Bog
cat 4-unitigger/asm.cga.0
Global Arrival Rate: 0.443659
There were 158,805,551 unitigs generated.
Unitig Length
Global Arrival Rate: 0.443659 # ??? <=> 200X cvg
100071 - 168549: 21
90845 - 99102: 15
80566 - 88867: 17
70006 - 79485: 39
60191 - 69891: 51
50210 - 59643: 98
40106 - 49917: 191
30015 - 39986: 448
20006 - 29992: 1068
10000 - 19995: 4187
9001 - 9999: 942
8001 - 8998: 1202
7000 - 7999: 1489
6000 - 6999: 1927
5000 - 5999: 2379
4000 - 4999: 3266
3000 - 3999: 4580
2000 - 2999: 6979
1000 - 1999: 9654
900 - 999: 1176
800 - 899: 1346
700 - 799: 1658
600 - 699: 2405
500 - 599: 4742
400 - 499: 13047
300 - 399: 26578
200 - 299: 361389
100 - 199: 135260255
90 - 99: 7874207
80 - 89: 7147630
70 - 79: 5128367
63 - 69: 2427507
138,219,089 out of 158,805,551 contain one of the frequent kmers
CGW
- Monitor cgw
ps -C cgw PID PPID %MEM RSZ %CPU STIME TIME CMD 8563 8560 95.2 251872528 88.2 13:24 01:47:56 /fs/szdevel/dpuiu/SourceForge/wgs-6.1/Linux-amd64/bin/cgw ...
top -b -p 8563 -d 10 | grep dpuiu > cgw.resource_usage.log
- Failure 1:
tail 7-0-CGW/cgw.out ... Processed 158,288,858 unitigs with 326,296,236 fragments #Bumble bee : Processed 61,930,044 unitigs with 301,738,113 fragments * Loaded dist s_2_3kb,1 (3000 +/- 300) * Loaded dist s_2_8kb,2 (8000 +/- 800) * Loaded dist s_3,3 (475 +/- 47.5) ... * Splitting chimeric input unitigs LIB 1 mu = 15.318100 sigma = 89.035478 LIB 2 mu = 8000.000000 sigma = 800.000000 LIB 3 mu = 337.817628 sigma = 26.699549 ... minLength = 460 minSplit = -429 Splitting unitig 47689 into as many as 3 unitigs at intervals: 22905,22906 .. Splitting unitig 158234882 into as many as 3 unitigs at intervals: 124,136 * BuildGraphEdgesDirectly
Fix (partial): add "-I" flag to cgw in runCA cat 7-0-CGW/cgw.out ... *** BuildGraphEdgesDirectly Operated on 171664374 fragments
- Failure 2:
tail 7-0-CGW/cgw.out **** Calling CheckEdgesAgainstOverlapper **** **** Survived CheckEdgesAgainstOverlapper with 0 failures**** * Allocating Contig Graph with 158289029 nodes and 14055921 edges Could not calloc memory (25326244640 * 1 bytes = 25326244640) cgw: AS_UTL_alloc.C:55: void* safe_calloc(size_t, size_t): Assertion `p != __null' failed. Fix : delete single fragment unitigs (tigStore) and the fragments asseociated with them (gkpStore)
158,288,858 unitigs total
154,631,861 unitigs to delete (single frg unitigs)
3,656,997 unitigs to keep
326,236,387 frg total
154,631,861 frg to delete (single frg unitigs)
171,604,526 frg to keep
Stats
. elem min q1 q2 q3 max mean n50 sum scf 11,768 109 4232 8819 22525 741575** 21676 51546 255094767 ctg 16,852 64 3501 7433 17418 317387** 15122 31217 254844680 ctg100+ 16,822 100 3514 7453 17442 317387 15149 31217 254842109 deg 3,388,183 64 125 138 168 4608 150 148 511490229 utg 3,657,090 64 124 138 169 168543 216 179 792973816 totalReads 326,236,387 usableReads 171,664,375 singletonReads 1
- Mate stats (%)
lib badLong badOuttie badSame bothDegen bothSurrogate diffScaffold good notMated oneDegen oneSurrogate 1 0.01 53.9 0 15.57 0.36 0.24 0.4 18.33 9.45 1.68 2 0 1.1 0 26.09 0.31 0.09 0.03 61.57 9.42 1.32 3 0.02 0 0 7.94 0.5 0.11 69.91 19.88 1.03 0.48 4 0.02 0 0 7.78 0.48 0.11 69 21.01 1.01 0.47 5 0.02 0 0 7.71 0.47 0.11 69.02 21.08 1 0.47 6 0.02 0 0 7.72 0.48 0.11 69.76 20.32 1 0.47 7 0.02 0 0 7.74 0.48 0.11 69.2 20.89 1 0.46
lib mates ULK CLK SLK 1 2494975 961180 521360 14 2 14560 14483 12375 4 3 13510875 2120862 1169185 2502 4 13100370 2048500 1128523 2392 5 12974843 2017970 1110081 2467 6 13304733 2059656 1125182 2481 7 12774596 1986181 1095046 2386
cat SLK.asm | grep ^mea | sed 's/mea://' | getSummary.pl -t SLK | pretty -int -o cat CLK.asm | grep ^mea | sed 's/mea://' | getSummary.pl -t CLK | pretty -int -o cat ULK.asm | grep ^mea | sed 's/mea://' | getSummary.pl -t ULK | pretty -int -o cat SCF.asm | grep mea | grep -v mea:0.000 | sed 's/mea://' | getSummary.pl -t CTP | pretty -int -o
. elem min q1 q2 q3 max mean n50 sum SLK 981 -53589 -484 -328 -183 4607 -1311 4607 -1286505 CLK 2714158 -488421 -42 34 73 7856 -682 7856 -1853032141 ULK 3576611 -188986 -71 24 69 7856 -338 7856 -1209800223 CTP 4987 -33482 -19 -6 15 10646 1 10646 8602
Location
mulberry:/scratch2/dpuiu/Megachile_rotundata/Assembly/wgs-noOBT/ (to be deleted) /fs/szattic-asmg5/Bees/Megachile_rotundata/Assembly/wgs-noOBT/ **
- Ftp
ftp://ftp.cbcb.umd.edu/pub/data/assembly/Megachile_rotundata.wgs.1/ /fs/ftp-cbcb/pub/data/assembly/Megachile_rotundata.wgs.1
- Try:
- bog
-b Break promisciuous unitigs at unitig intersection points => delete -m 7 Break a unitig if a region has more than 7 bad mates => increase to 1000
- cgw :
-m <min> Number of mate samples to recompute an insert size, default is 100 => increase to ?
SOAPdenovo (Tanja)
cat *.ContigIndex | grep -v ^E | grep -v ^i | count.pl -i 1 | getSummary.pl -j 1 -t "contigs" cat *.ContigIndex | grep -v ^E | grep -v ^i | count.pl -i 1 | getSummary.pl -j 1 -min 100 -t "contigs(>100bp)" grep "^>" *.scaf | getSummary.pl -i 2 -t scaf
- Stats
. elem min q1 q2 q3 max mean n50 sum scaf 7863 102 903 3272 17692 2,338,728** 37825 240,706 297423517 # N50 for Bee was 1.17M contigs 9742349 31 32 33 37 114,832 60 44 585430821 contigs(>100bp) 177327 100 131 261 1398 114,832 1333 3897** 236496823 # N50 for Bee was 7K
- Location
/fs/szattic-asmg5/Bees/Megachile_rotundata/Assembly/assembly5kbForAll
SOAPdenovo (Daniela)
Stats
. elem min q1 q2 q3 max mean n50 sum scaff 25,119 351 1896 4444 10914 1,102,803 11041 26876 277,338,897 scaff(all) 51,551 100 121 489 4320 1,102,830 5516 25940 284,368,749 scaffContigs(all) 263,977 33 78 149 878 121,523 914 3273 241,406,149 # <= scafffasta2fasta.pl contigs(all) 6,917,796 31 32 34 40 121,554 70 73 487,401,812 contigs(>100bp) 210,666 100 124 222 1174 121,554 1108 3138 233,563,401 reads 340,437,486 readsOnContigs 171,212,613
Alignments
- Align reads to the scaffolds
soap2-index asm.K31.scafSeq
mkdir soap2-index
mv asm.K31.scafSeq.index.* soap2-index/
soap2 -D soap2-index/asm.K31.scafSeq.index -a s_2_1_1.1kb_sequence.txt -b s_2_2_1.1kb_sequence.txt -l 32 -p 16 -v 2 -m 800 -x 1400 -o s_2_1.1kb.mated.soap2 -2 s_2_1.1kb.single.soap2
soap2 -D soap2-index/asm.K31.scafSeq.index -a s_2_1_3kb_sequence.txt -b s_2_2_3kb_sequence.txt -l 32 -p 16 -v 2 -m 2000 -x 4000 -o s_2_3kb.mated.soap2 -2 s_2_3kb.single.soap2 -R
soap2 -D soap2-index/asm.K31.scafSeq.index -a s_2_1_5kb_sequence.txt -b s_2_2_5kb_sequence.txt -l 32 -p 16 -v 2 -m 4000 -x 6000 -o s_2_5kb.mated.soap2 -2 s_2_5kb.single.soap2 -R
soap2 -D soap2-index/asm.K31.scafSeq.index -a s_2_1_8kb_sequence.txt -b s_2_2_8kb_sequence.txt -l 32 -p 16 -v 2 -m 6000 -x 10000 -o s_2_8kb.mated.soap2 -2 s_2_8kb.single.soap2 -R
soap2 -D soap2-index/asm.K31.scafSeq.index -a s_3_1_sequence.txt -b s_3_2_sequence.txt -l 32 -p 16 -v 2 -m 200 -x 400 -o s_3.mated.soap2 -2 s_3.single.soap2
...
mates mated single single.diffScaff single.sameScaff
s_2_1.1kb 32,634,858 1,466,112 8,014,900 131,084 585,668
s_2_3kb 21,563,283 1,545,114 8,449,321 341,974 1,203,570
s_2_3kb.trim64 21,563,283 4,291,750* 12,156,958 1,484,498* 3,457,492
s_2_3kb.filter 4,823,235 3,730 3,618,426 38,562 3,017,172
s_2_5kb 36,218,589 5,639,332 44,533,553 4,784,348 30,621,038
s_2_8kb 198,377 1,068 32,280 1,168 2,842
s_2_8kb.trim64 198,377 5,508* 48,877 4,258* 3,532
s_2_8kb.filter 111,267 20 33,819 372 27,300
s_3 35,548,153 15,521,020 4,589,276 37,564 651,924
Location
mulberry:/scratch2/dpuiu/Megachile_rotundata/Assembly/SOAPdenovo/
SOAPdenovo ; partial : s_[34567] ; no repeats **
- Slightly better results than we got when the s_2_?kb libs were used
Stats
. elem min q1 q2 q3 max mean n50 sum scaff 24,602 333 1724 4380 11049 1,103,462 11135 27887 273963709 scaff(all) 40,883 100 146 1366 5960 1,103,447 6833 27088 279355387 scaffContigs(all) 231,559 33 75 153 982 148,167 1039 3950 240752397 contigs(all) 2,515,516 31 33 36 52 148,198 131 1880 330512911 contigs(100bp+) 184,395 100 127 235 1316 148,198** 1263 3730 232932308
Location
ginkgo: /scratch1/dpuiu/Megachile_rotundata/Assembly/SOAPdenovo-partial.3.noRepeats/
Aligning long inserts to this assembly =
- Should trim the linker (mostly at the beginning of the reads )
show-coords -H 1000_12.filter-1.delta | sort -k19 | p '$F[18]=~s/[12]$//; print $p,$_,"\n" if($P[18] eq $F[18] and $P[17] ne $F[17]); $p=$_; @P=@F;' | pretty ... cat *-1000*filter*delta | show-coords.pl | sort -k19 | ...
417 469 | 2 54 | 53 53 | 98.11 | 2148 54 | 2.47 98.15 | scaffold16793 HWI-EAS385_0062:2:1:10306:11665#CAGATC/1 [CONTAINS] 10634 10719 | 39 124 | 86 86 | 96.51 | 10732 124 | 0.8 69.35 | scaffold21864 HWI-EAS385_0062:2:1:10306:11665#CAGATC/2 . 1 38 | 40 3 | 38 38 | 100 | 42 124 | 90.48 30.65 | linker.rev HWI-EAS385_0062:2:1:10306:11665#CAGATC/2 .
1 38 | 40 3 | 38 38 | 100 | 42 124 | 90.48 30.65 | linker.fwd HWI-EAS385_0062:2:1:10759:13923#CAGATC/1 . 1 82 | 41 122 | 82 82 | 95.12 | 7015 124 | 1.17 66.13 | scaffold5186 HWI-EAS385_0062:2:1:10759:13923#CAGATC/1 . 5293 5416 | 1 124 | 124 124 | 94.35 | 7427 124 | 1.67 100 | scaffold7361 HWI-EAS385_0062:2:1:10759:13923#CAGATC/2 [CONTAINS]
SOAPdenovo ; s_2_3kb & s_ 2_8kb soap-aligned & trim64; s_2_1.1k & s_2_1.4k ; s_[34567] no repeat **
Stats
. elem min q1 q2 q3 max mean n50 sum scaf 6242 228 640 2061 10681 3,022,211 42964 456,467 268,183,797 scafSeq 17702 100 119 178 830 2,999,853 15136 452,601 267,954,678 contigs 4138734 32 32 34 41 111,437 94 1286 389,303,351 contigs100+ 181683 100 131 252 1385 111,437 1307 3789 237,563,819 scafSeqContigs 124749 2 90 466 2237 123,623 1970 5,800 245,850,885 scafSeqContigsClosed 25288 2 130 382 5576 626,941 10218 61,363 258,400,185 # after running GapCloser
- GC contig cvg bias
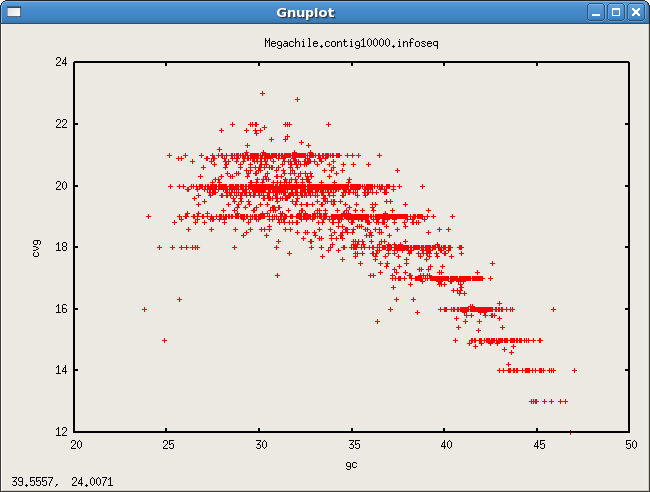
Scaffold links
perl ~/bin/remapSOAPscafId.pl *.scaf -p 5 | p 'print $_ unless($F[-2]=~/DOWN/ and $F[-1]=~/UP/);' | more ... >2 76 50030 2.2 206 58 #DOWN #UP 543.152:5:1248 2.4 611 1458 #DOWN #UP 543.152:26:1477 2.24 18951 911 #DOWN 2268.26:31:139 #UP 2.25 20219 2382 #DOWN #UP 2268.26:28:212 2.40 34053 150 #DOWN 1363.46:10:1041 #UP 2.42 34434 76 #DOWN 1363.46:6:804 #UP 2.43 34500 70 #DOWN 1363.46:6:682 #UP 2.60 41566 200 #DOWN 808.70:8:219 #UP 2.62 42030 1405 #DOWN #UP 808.70:9:82 .. >543 153 191782 543.63 108072 1765 #DOWN 158.305:5:198 #UP 543.66 110202 77 #DOWN #UP 158.305:6:185 543.98 163144 810 #DOWN 1285.8:25:206 #UP 543.99 164653 393 #DOWN #UP 91.485:6:197 543.121 176938 49 #DOWN #UP 985.12:24:88 543.125 178897 102 #DOWN 234.161:5:275 #UP 543.128 179479 63 #DOWN 220.220:11:214 1951.10:14:284 576.114:16:256 #UP 576.116:8:474 234.161:8:178 543.152 188359 3153 #DOWN 2.4:26:1477 2.2:5:1248 #UP ...
SOAPdenovo vs CA
Aligns contigs 200+bp with nucmer.amos
set REFN=`grep -c ">" ref.seq` @ REFN/=20 nucmer.amos -D REF=ref -D QRY=qry -D REFN=$REFN ref-qry
. elem min q1 q2 q3 max mean n50 sum ref(CA) 16507 200 3678 7660 17785 317387 15436 31240 254794314 qry(SOAPdenovo) 100877 200 499 1224 2328 111437 2248 4103 226807646
cat ref-qry.delta | grep "^>" | sed 's/>//' | awk '{print $1,$3}' | sort -u | getSummary.pl -i 1
. elem min q1 q2 q3 max mean n50 sum
ref-hits 16195 200 3816 7848 18116 317387 15711 31309 254444709
qry-hits 95343 200 610 1307 2427 111437 2356 4171 224616404
. elem min q1 q2 q3 max mean n50 sum ref-miss 312 200 244 483 1575 13984 1121 2262 349605 qry-miss 5534 200 235 298 442 5658 396 421 2191242
Location
mulberry: /scratch2/dpuiu/Megachile_rotundata/Assembly/SOAPdenovo.noRepeats.trim64/ (to be deleted) /fs/szattic-asmg5/Bees/Megachile_rotundata/Assembly/SOAPdenovo.noRepeats.trim64/ ** /fs/ftp-cbcb/pub/data/assembly/Megachile_rotundata.SOAPdenovo.2/ => ftp://ftp.cbcb.umd.edu/pub/data/assembly/Megachile_rotundata.SOAPdenovo.2/
SOAPdenovo ; s_2_3kb & s_ 2_8kb soap-aligned & trim64; s_2_1.1k,s_2_1.4k,s_[34567] no repeat & trim64
- Trimming all reads to the first 64bp does not change the results much (actually getting a little worse)
Stats
. elem min q1 q2 q3 max mean n50 sum scaf 7615 441 896 2238 11462 2394735 37960 326734 289063269 scafSeq 23906 100 118 170 914 2396948 12271 324184 293340377 contigs 1738246 32 33 37 61 122972 174 2037 302039034 contigs100+ 214540 100 135 264 1263 122972 1110 2941 238063650
SOAPdenovo K=31 (new data)
- Location
/scratch1/dpuiu/Megachile_rotundata/Assembly/SOAPdenovo/K31 ftp://ftp.cbcb.umd.edu/pub/data/assembly/Megachile_rotundata.SOAPdenovo.2011-03/
- Assembly stats:
. elem min q1 q2 q3 max mean n50 sum scf 16115 100 111 134 264 4173260 17307 1288790 278898652 scfLen 16115 100 111 134 208 3855147 14496 950327 233605903 ctg 8829648 32 32 34 39 121554 62 3201 553630969
scf2 16115 100 111 134 260 4033560 16539 1067152 266528571 scf2Len 16115 100 111 134 259 4004254 16236 1071079 261641263 ctg2 26490 3 120 243 5253 520023 9877 64739 261641355
cat asm.K31.peGrads | tail -6 | p 'print $F[0], " ", $F[1]-$P[1],"\n"; @P=@F' | pretty 350 330665408 1100 54707484 1400 86601708 3000 22148926 5300 37909358 8000 30203916
cat asm.K31.links | awk '{print $5}' | uniq -c | awk '{print $2,$1}'
350 7375561
1100 579996
1400 604951
3000 340192
5300 669339
8000 184868
7375561 350 579996 1100 604951 1400 340192 3000 669339 5300 184868 8000
SOAPdenovo K=47 (new data) **
. elem min q1 q2 q3 max mean n50 sum scaf 3495 259 408 757 4763 6,173,378 82382 2,124,089 287,925,734 scafSeq 31774 100 110 128 172 6,174,792 9215 2,124,853* 292,784,153 scafSeq2 31774 100 110 128 172 5,876,085 8689 1,814,396 276,082,351 contigs 10217806 48 48 50 56 175,671 77 4,701 792,925,336 contigs100+ 224315 100 126 177 933 175,671 1134 4,701 254,309,160 scafSeqContigsClosed 43232 2 114 147 740 479,105 6228 63,194* 269,243,097
Location:
/fs/szattic-asmg5/Bees/Megachile_rotundata/Assembly/SOAPdenovo.10libs.K47
SOAPdenovo vs CA
elem min q1 q2 q3 max mean n50 sum CA 16848 64 3501 7433 17428 317387 15124 31240 254808119 SO 43232 2 114 147 740 479105 6228 63194 269243097
CA.no_hits 315 64 124 150 188 1353 168 167 52986 SO.no_hits 29679 2 109 123 151 5766 185 158 5489188
Allpaths-LG (experiment)
- Shred SOAPdenovo K=47 contigs >=180bp ; use them as fragment library
. elem min q1 q2 q3 max mean n50 sum contig.180+ 110580 180 264 964 2321 175671 2166 4701 239535581
- Libraries used: (in_libs.csv)
library_name, project_name, organism_name, type, paired, frag_size, frag_stddev, insert_size, insert_stddev, read_orientation, genomic_start, genomic_end #mates frag, genome, genome, fragment, 1, 180, 20, , , inward, 0, 0 22,024,446 # originally 176,833,196 fragments insert_size=475bp s_2_1.1kb, genome, genome, jumping, 1, , , 1100, 110, inward, 0, 0 32,634,858 s_2_1.4kb, genome, genome, jumping, 1, , , 1400, 140, inward, 0, 0 50,861,645 s_7_5kb, genome, genome, jumping, 1, , , 5300, 530, outward, 0, 0 29,111,787 s_7_8kb, genome, genome, jumping, 1, , , 8000, 800, outward, 1, 37 25,328,718
type #mates.original #mates.corrected frag 22,024,446 21,925,564 jumping 137,937,008 5,465,197
- Assembly stats:
. elem min q1 q2 q3 max mean n50 sum scf 1964 1 1187 1550 6679 6,117,056 128,398 1,806,225 252,173,219 ctg 36959 1 1836 3140 6343 278,684 6,061 8,784 224,007,758
Genbank submission
- Information
Project Type: Single Species Project
Contacts: Daniela Puiu dpuiu@umiacs.umd.edu, Steven Salzberg salzberg@umiacs.umd.edu
Submitting Organization: University of Illinois & University of Maryland
Sequencing Center: Keck Center for Comparative and Functional Genomics, University of Illinois BCUI Biotechnology Center, Univ. Illinois (BCUI)
Consortium Name: Alfalfa Leafcutter Bee Genome Consortium
Organism Name: Megachile rotundata
Strain/isolate/breed: North American commercial strain
Locus Tag Prefix: MROT #3+ letters
Source of DNA used for sequencing: whole body, haploid brother males
Sequencing Method: wgs
Sequencing Technology: Illumina
Estimated Genome Size: 250Mb # the haploid genome size
Brief description of the importance: Megachile rotundata, alfalfa leafcutting bee, is a solitary bee species. It is the #3 agricultural pollinator in the United States and is commercially managed for alfalfa seed production.
Comments to the staff:
DNA
whole genome sequencing
single genome;
no annotation
assembly name MROT_1.0
assembly method: SOAPdenovo Assembler v_1.05
plan to update: ?
expect to release : ?
strain information to be submitted soon
genome coverage: 300x
sequencing technology: Illumina GA IIx ??
Author list:
Gene E. Robinson,
Hugh M. Robertson,
Matthew E. Hudson,
Kim Walden,
Brielle J. Fischman,
Theresa Pitts-Singer,
Rosalind James
Steven Salzberg,
Daniela Puiu,
Tanja Magoc,
David Kelley,
Aleksey Zimin ,
- Best assembly : SOAPdenovo K=47
/fs/szattic-asmg5/Bees/Megachile_rotundata/Assembly/SOAPdenovo.10libs.K47/genbank[1-3]
- Sequence length statistics [1]:
number min max mean n50 sum scaffolds 31774 100 5876085 8689 1814396 276082351 contigs 42781 100 479105 6293 69010 269224383
- Removed 6 contaminants & scaff < 200bp [2]
. elem min max mean n50 sum scaffolds 6367 200 5876085 42857 1699680 272873468 contigs 17374 100 479105 15311 64153 266015500
- NCBI comments: There are 668 contigs that are <200bp remaining. Some of these are internal components of scaffolds, but many of these are at the ends of scaffolds so should be removed. For example, 249 of them are the first or component of a scaffold or the only component of a singleton scaffold (list.ShortTerminalContigs). And some of them are the only components of a multi-component scaffold, eg these two scaffolds are made entirely of short contigs ...
. elem min max mean n50 sum scaffolds 6266 200 5876085 43514 1814396 272660569 # 101 scaffolds deleted contigs 16706 200 479105 15917 69010 265916502 # 668 contigs deleted
- GPID Organism name Accession
----------------------------------------------- 66515 Megachile rotundata AFJA00000000
Please cite the accession number as usual:
This Whole Genome Shotgun project has been deposited at DDBJ/EMBL/GenBank under the accession AFJA00000000. The version described in this paper is the first version, AFJA01000000.