Data
Original Traces
- 8 pairs of data files (paired ends)
cat trace.count | grep _1_ | sed 's/_sequence.txt//' | perl -ane 'print " ",$F[1],"\t",$F[0]/4,"\t",$F[0]/2,"\n";'
lib insert mates reads readLen ~coverage(500M genome)
s_2_3kbp 3000 21,563,283 43,126,566 124 11
s_2_8kbp 8000 198377 396,754 124 0.1
s_3 475 35548153 71,096,306 124 18
s_4 475 35471044 70,942,088 124 18
s_5 475 35616846 71,233,692 124 18
s_6 475 35303840 70,607,680 124 18
s_7 475 34893313 69,786,626 124 18
total . 198,594,856 397,189,712 128 98*
Corrected Traces
lib insert mates reads repeatReads
s_2_3kb 3000 4,823,235 9,646,470 4,349,208 (45%)
s_2_8kb 8000 111,267 222,534 167,246 (75%)
s_3 475 33,024,597 66,049,194 35,777,342 (54%)
s_4 475 33,237,593 66,475,186
s_5 475 33,150,790 66,301,580
s_6 475 33,223,371 66,446,742
s_7 475 32,647,890 65,295,780
total . 170,218,743 340,437,486
- repeatReads:
- at least one of the mate contains a perfect match of one of the 15 frequent 22mers listed below
- 32.5%GC in repeatREads vs ~ 35.5%GC in uniqueReads
Adaptors
>circularizarion
CGTAATAACTTCGTATAGCATACATTATACGAAGTTATACGA
>circularizarion.revcomp
TCGTATAACTTCGTATAATGTATGCTATACGAAGTTATTACG
Frequent kmers
- 22mers which seem to appear in tandem
1 AATCATACAATCACAATCATAC|GTATGATTGTGATTGTATGATT : AATCATAC AATCAC|GTGATTGTATGATT #14mer
2 CAATCACAATCATACAATCACA|TGTGATTGTATGATTGTGATTG
3 AATAATATGAGTTAGATTGATA|TATCAATCTAACTCATATTATT
4 ATATAAGCATAATATGGCTAAT|ATTAGCCATATTATGCTTATAT
5 AGTAATTGTCGTTCTATCGATC|GATCGATAGAACGACAATTACT
6 AGACAGAGACAGAGACAGAGAC|GTCTCTGTCTCTGTCTCTGTCT
7 TCACACAATCACAATCACACAA|TTGTGTGATTGTGATTGTGTGA
8 CACACAATCACACAATCACACA|TGTGTGATTGTGTGATTGTGTG
9 ATTACTCTTATTATTATCAATC|GATTGATAATAATAAGAGTAAT
10 CTGTCTCTGTCTGTCTCTGTCT|AGACAGAGACAGACAGAGACAG
11 ACAATTACTATACTTATTACTC|GAGTAATAAGTATAGTAATTGT
12 CACAATCACGATCACACAATCA|TGATTGTGTGATCGTGATTGTG
13 AACCTAACCTAACCTAACCTAA|TTAGGTTAGGTTAGGTTAGGTT
14 CAGCGGATATGTGCGAATTAGA|TCTAATTCGCACATATCCGCTG
15 CTGAGCACAATTCAACACCACA|TGTGGTGTTGAATTGTGCTCAG
Location
/fs/szattic-asmg5/Bees/Megachile_rotundata/error_correction/large_libs/s_?_?_?kb.sequence.cor.all.txt
/fs/szattic-asmg5/Bees/Megachile_rotundata/error_free/s_?_?_sequence.cor.txt
/fs/szattic-asmg5/Bees/Megachile_rotundata/frg/ # frg files to assemble
Assemblies
- CA Version: 6.1 (09/01/2010) /fs/szdevel/dpuiu/SourceForge/wgs-6.1/Linux-amd64/bin/runCA
- SOAP version 1.04: /nfshomes/dpuiu/szdevel/SOAPdenovo_Release1.04/
CA noOBT partial
- Data : 3 libs : ~ 16X cvg
Gatekeeper
LibraryName numActiveFRG numDeletedFRG numMatedFRG readLength clearLength
GLOBAL 72,995,448 0 70632632 8307194830 8278360381
LegacyUnmatedReads 0 0 0 0 0
s_2_3kb 9,166,343 0 8736228 942501164 914798596
s_2_8kb 210,266 0 199620 21669112 20742291
s_3 63,618,839 0 61696784 7343024554 7342819494
UID IID mateUID mateIID libUID libIID isDel isNonRandom Orient Length clrBeginLATEST clrEndLATEST
110000000001 1 120000000001 2 s_2_3kb 1 0 0 I 75 0 75
120000000001 2 110000000001 1 s_2_3kb 1 0 0 I 123 0 123
110000000003 3 120000000003 4 s_2_3kb 1 0 0 I 90 0 90
120000000003 4 110000000003 3 s_2_3kb 1 0 0 I 123 40 123
...
110009166343 9166343 0 0 s_2_3kb 1 0 0 U 76 11 76
210009166344 9166344 220009166344 9166345 s_2_8kb 2 0 0 I 123 21 123
...
210009376609 9376609 0 0 s_2_8kb 2 0 0 U 88 0 88
320009376610 9376610 0 0 s_3 3 0 0 U 72 0 72
...
310072995448 72995448 0 0 s_3 3 0 0 U 68 0 68
gatekeeper -b 1 -e 2 -dumpfastaseq asm.gkpStore/ | more
>110000000001,1 mate=120000000001,2 lib=s_2_3kb,1 clr=LATEST,0,75 deleted=0
TAATAATATGCGTTAGAGTGATAATAATTGGAGTAATGAGTATAGCAATTGTCGTCCTGTCGATCATATCTGCAT
>120000000001,2 mate=110000000001,1 lib=s_2_3kb,1 clr=LATEST,0,123 deleted=0
CCATATTATGCTTATATGATCGATAGAACGACAATTACTATACTTATTATTCTTATTATTATCAATCTCACTCATATTATTAACCATATTATGCAGACATGATCGATAGAACGGCAATTACTA
head -2 s_2_3kb_?.filter.seq
>HWI-EAS385_0062:2:1:1036:15608#GCCAAT/1
TAATAATATGCGTTAGAGTGATAATAATTGGAGTAATGAGTATAGCAATTGTCGTCCTGTCGATCATATCTGCAT
>HWI-EAS385_0062:2:1:1036:15608#GCCAAT/2
CCATATTATGCTTATATGATCGATAGAACGACAATTACTATACTTATTATTCTTATTATTATCAATCTCACTCATATTATTAACCATATTATGCAGACATGATCGATAGAACGGCAATTACTA
Stats
. elem min q1 q2 q3 max mean n50 sum #repeats comments
scf 20,827 122 3228 6374 13700 202495 11508 20462 239696810 SOAPdenovo: max=1102803 , N50=26876
ctg 37,494 65 2185 3998 7706 191323* 6380 10151* 239226293 206 SOAPdenovo: max=121554 , N50=3138
deg 1,136,469 64 123 143 184 5031 160 164 181954480 807132
utg 1,437,146 64 123 143 195 67048 308 870 443759899
readsTotal 72,995,448
readsInContigs 27,837,956
readsInDegenerates 9,627,122
singletons 34,881,692 (47%) 31,987,184
readsWithOuttieMate 3,028,956(4.15%) ???
Issues
- lib s_2_* orientation ???
cat asm.posmap.frags | grep placed | p '/(\w)/; print "$1 $F[4]\n";' | count2col.pl | pretty -o
. badLong badOuttie badSame bothDegen bothSurrogate diffScaffold good notMated oneChaff oneDegen oneSurrogate
1 534 2,998,286 458 1614846 9892 21872 27308 267328 979980 760044 65268
2 4 26,864 10 38636 114 294 178 5044 35465 7848 1022
3 11072 3,806 1104 2369982 61236 53370 23058022 1208112 3967689 371538 87260
cat asm.posmap.frags | grep -v placed | p '/(\w)/; print "$1 $F[4]\n";' | count2col.pl | pretty -o
. bothChaff notMated oneChaff
1 1,277,760 162,787 979,980
2 53,588 5,602 35,465
3 27,684,878 713,943 3,967,689
Location
ginkgo:/scratch1/dpuiu/Megachile_rotundata/Assembly/wgs-noOBT-partial/
Moved from
mulberry:/scratch2/dpuiu/Megachile_rotundata/Assembly/wgs-noOBT-partial/
CA noOBT
- Data : 7 libs : ~ 74X cvg
Gatekeeper
LibraryName numActiveFRG numDelFRG numMatedFRG readLength clearLength #repeats
GLOBAL 326,236,387 0 315518526 37451489553 37418130441
LegacyUnmatedReads 0 0 0 0 0
s_2_3kb 9107424 0 9107424 942165284 910444046 #
s_2_8kb 209336 0 209336 21814418 20787384 #
s_3 63618839 0 61696784 7343024554 7342819494 #
s_4 63544688 0 61255960 7291557748 7291478152 #
s_5 63370860 0 61084368 7271218123 7271051639 #
s_6 63780887 0 61685156 7359094156 7359012512 #
s_7 62604353 0 60479498 7222615270 7222537214 #
Meryl
meryl -Dh -s 0-mercounts/asm-C-ms22-cm0
Found 30570218845 mers.
Found 271464470 distinct mers.
Found 11164787 unique mers.
Largest mercount is 87984949; 1896 mers are too big for histogram.
1 11164787 0.0411 0.0004
2 9376915 0.0757 0.0010
3 3714582 0.0894 0.0013
...
54 5344148 0.6573 0.1788
...
fasta2tab.pl 0-mercounts/asm.nmers.ovl.fasta | sort -n -r | head -5
87,908,217 AATCATACAATCACAATCATAC
84,450,288 CAATCATACAATCACAATCATA
...
74,975,282 AATAATATGAGTTAGATTGATA
egrep -c 'AATCATACAATCACAATCATAC|GTATGATTGTGATTGTATGATT' *fastq *txt > egrep.count
mulberry:/scratch2/dpuiu/Megachile_rotundata/Data/error_free/egrep.count
meryl -Dh -s 0-mercounts/asm-C-ms15-cm0 | head
Found 32850820919 mers.
Found 142500876 distinct mers.
Found 2381895 unique mers.
Largest mercount is 125816941; 2023 mers are too big for histogram.
1 2381895 0.0167 0.0001
2 2325770 0.0330 0.0002
3 708786 0.0380 0.0003
...
54 1851586 0.4894 0.0671
...
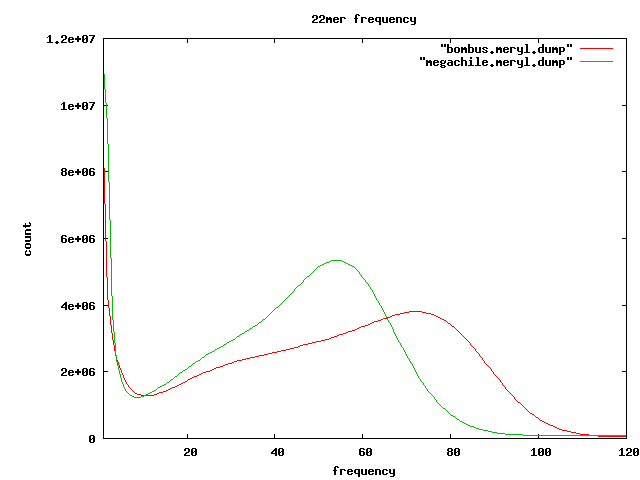
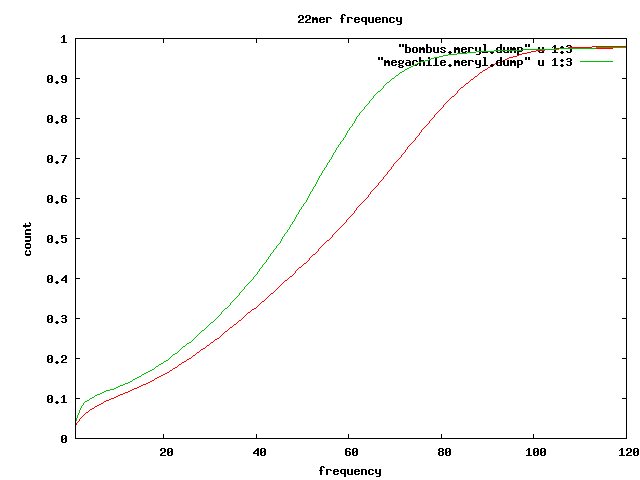
Overlap
cat 1-overlapper/ovlopts.pl | grep ^\"h | wc -l
924
- Failures: 709 jobs failed; runCA 6.1 could not restart overlap properly !!!
cat 1-overlap/overlap*out | grep "^Could not" | sort -u
Could not malloc memory (1305184948 bytes)
cat 1-overlapper/*pl | grep ^\"0 | sed 's/"//' | sed 's/\",//' >! 1-overlapper/ovlopts.pl.0
cat 1-overlapper/*pl | grep ^\"h | sed 's/"//' | sed 's/\",//' >! 1-overlapper/ovlopts.pl.h
cat 1-overlapper/*pl | grep ^\"-h | sed 's/"//' | sed 's/\",//' >! 1-overlapper/ovlopts.pl.-h
paste 1-overlapper/ovlopts.pl.* | p 'print "overlap -M 8GB --hashload 0.8 -t 1 -h $F[3] -r $F[5] -k 22 -k \
./0-mercounts/asm.nmers.ovl.fasta -o ./1-overlapper/$F[0]/$F[1].ovb.gz ./asm.gkpStore > \
./1-overlapper/overlap.0$..out \n";' | tail -709 > overlap.sh
overlapStore -d asm.ovlStore | awk '{print $1}' | uniq -c | awk '{print $1}' | count.pl | getSummary.pl -i 0 -j 1
overlapStats -G asm.gkpStore -O asm.ovlStore -o asm
Location
mulberry:/scratch2/dpuiu/Megachile_rotundata/Assembly/wgs-noOBT
SOAPdenovo (Tanja)
cat *.ContigIndex | grep -v ^E | grep -v ^i | count.pl -i 1 | getSummary.pl -j 1 -t "contigs"
cat *.ContigIndex | grep -v ^E | grep -v ^i | count.pl -i 1 | getSummary.pl -j 1 -min 100 -t "contigs(>100bp)"
grep "^>" *.scaf | getSummary.pl -i 2 -t scaf
. elem min q1 q2 q3 max mean n50 sum
contigs 9742349 31 32 33 37 114832 60 44 585430821
contigs(>100bp) 177327 100 131 261 1398 114832 1333 3897 236496823 # N50 for Bee was 7K
scaf 7863 102 903 3272 17692 2338728 37825 240706 297423517 # N50 for Bee was 1.17M
/fs/szattic-asmg5/Bees/Megachile_rotundata/Assembly/assembly5kbForAll
SOAPdenovo (Daniela)
cat asm.K31.contig | grep "^>" | awk '{print $3}' | uniq -c | awk '{print $2,$1}' > asm.K31.contigLen.count
. elem min q1 q2 q3 max mean n50 sum
contigs(all) 6,917,796 31 32 34 40 121554 70 73 487,401,812
contigs(>100bp) 210,666 100 124 222 1174 121554* 1108 3138* 233,563,401
scaff 25,119 351 1896 4444 10914 1102803 11041 26876 277,338,897
reads 340,437,486
readsOnContigs 171,212,613
mulberry:/scratch2/dpuiu/Megachile_rotundata/Assembly/SOAPdenovo-redo

