Trace formatting: Difference between revisions
| (168 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
= Articles = | |||
* [http://www.sciencedirect.com/science?_ob=ArticleURL&_udi=B6TCY-4RTCPK7-2&_user=961305&_rdoc=1&_fmt=&_orig=search&_sort=d&view=c&_acct=C000049425&_version=1&_urlVersion=0&_userid=961305&md5=866485013aea95d971ee7d3b4e5e7244 The impact of next-generation sequencing technology on genetics] Elaine Mardis Trends in Genetics 2008 | |||
* [http://www.in-sequence.com/issues/1_9/features/138789-1.html As Users Demand Paired-End Sequencing, 454, Illumina, and ABI Work On New Kits] | |||
* [http://www.ebi.ac.uk/help/formats.html Sequence Format Descriptions(EMBL)] | |||
* [http://minds.nuim.ie/~tkeane/publications/ismb2007Poster.pdf ismb2007Poster.pdf] | |||
* [http://www.sb-roscoff.fr/SG/IMG/pdf/Smith_Rennes_2007.pdf Smith_Rennes_2007.pdf] | |||
* [http://www.illumina.com/downloads/GenomeAnalyzer_SpecSheet.pdf GenomeAnalyzer_SpecSheet.pdf] | |||
* [http://www.genomeweb.com/pdfs/GTechSeqGuide1208.pdf Assembly and Alignment Algorithms for Next-Gen Sequence Data] | |||
* [http://genome.cshlp.org/content/18/12/2024 Sequencing of natural strains of Arabidopsis thaliana with short reads] (Illumina) | |||
* [http://genomebiology.com/2007/8/7/R143 Accuracy and quality of massively parallel DNA pyrosequencing] | |||
* [http://jeb.biologists.org/cgi/reprint/210/9/1518 Advanced sequencing technologies and their wider impact in microbiology] | |||
* [http://www.botany.uga.edu/~suew/DNA_Seq_newGR06.pdf Emerging technologies in DNA sequencing] | |||
* [http://www-hto.usc.edu/papers/msw_papers/msw-081.pdf Lander Waterman] | |||
* [http://www.sciencedirect.com/science?_ob=ArticleURL&_udi=B6VS0-4M4TNKN-1&_user=961305&_rdoc=1&_fmt=&_orig=search&_sort=d&view=c&_acct=C000049425&_version=1&_urlVersion=0&_userid=961305&md5=2d1969432fa6f4f1c3b5d23060d43fad Whole-genome re-sequencing ] | |||
* [http://www.pubmedcentral.nih.gov/picrender.fcgi?artid=1871613&blobtype=pdf Whole-Genome Sequencing and Assembly with High-Throughput, Short-Read Technologies] | |||
* [http://seqanswers.com/ seqanswers] | |||
* [http://www.politigenomics.com/next-generation-sequencing-informatics politigenomics table] | |||
* [http://www.valexllc.com/products.html Valex & SRA] | |||
* [http://bioinformatics.oxfordjournals.org/cgi/reprint/25/4/429.pdf BIOINFORMATICS FOR NEXT GENERATION SEQUENCING] | |||
* [http://www.genomeweb.com/short-read-headache The Short Read Headache] | |||
* [http://www.ncbi.nlm.nih.gov/pubmed/18262675?ordinalpos=11&itool=EntrezSystem2.PEntrez.Pubmed.Pubmed_ResultsPanel.Pubmed_DefaultReportPanel.Pubmed_RVDocSum The impact of next-generation sequencing technology on genetics] | |||
* [http://www.ncbi.nlm.nih.gov/pubmed/18576944?ordinalpos=7&itool=EntrezSystem2.PEntrez.Pubmed.Pubmed_ResultsPanel.Pubmed_DefaultReportPanel.Pubmed_RVDocSum Next-generation DNA sequencing methods.] | |||
* [http://www.ncbi.nlm.nih.gov/pubmed/19133817?ordinalpos=1&itool=EntrezSystem2.PEntrez.Pubmed.Pubmed_ResultsPanel.Pubmed_DefaultReportPanel.Pubmed_RVDocSum Short-Read Sequencing Technologies for Transcriptional Analyses.] | |||
* [http://www.oxfordjournals.org/our_journals/bioinformatics/nextgenerationsequencing.html Next Generation Sequencing - Bioinformatics Journal - Virtual Edition] | |||
* [http://www.sciencemag.org/products/lst_20090410.dtl Sanger Who Science April 2009] | |||
* [http://genome.cshlp.org/content/early/2011/07/18/gr.123638.111.full.pdf+html?rss=1 Accurate and comprehensive sequencing of personal genomes] Genome Research July 2011 | |||
= Cleaning = | |||
* run dust on Ecoli & UniVector to mask low complexity seqs | |||
* screen against Ecoli & UniVector database | |||
= Technologies = | |||
Latest Technology Summary | |||
Technology [http://www.454.com/ 454] [http://www.illumina.com/ Illumina] [http://www3.appliedbiosystems.com/AB_Home/applicationstechnologies/SOLiDSystemSequencing/index.htm Solid] | |||
seq-by-synthesis seq-by-synthesis ABI | |||
Company 454(Roche) Illumina | |||
Location Brandford,CT SanDiego,CA | |||
Latest GS FLX, Titanium reagents Genome Analyzer II SOLID 3 | |||
Throughput 500M/run 1G/run 20G/run | |||
RunTime 10hr 3days | |||
ReadLen 500bp 36 35 | |||
InsertLen 3K 100-200bp 600-10K | |||
Accuracy 99.94% | |||
Q20(99%accuracy) 400bp 34bp | |||
Cost $3K/run 60K/3G | |||
$400/4M bacterial genome(25-30X) | |||
Problems homopolimers | |||
== Sanger == | == Sanger == | ||
== 454 | == 454 == | ||
== | * Pyrosequencing | ||
* [http://en.wikipedia.org/wiki/454_Life_Sciences 454_Life_Sciences wikipedia] | |||
* [http://www.in-sequence.com/issues/1_37/features/142515-1.html In-sequenece article] | |||
* [http://www.454.com/downloads/protocols/1_paired_end.pdf 1_paired_end.pdf] | |||
* [http://www.nature.com/nmeth/journal/v5/n5/full/nmeth.f.212.html] | |||
* [http://arjournals.annualreviews.org/doi/abs/10.1146/annurev.genom.9.081307.164359] | |||
Anomalies: | |||
* homopolymer lengths can be shorter than real | |||
* substitutions less likely than in traditional methodssingle base insertions | |||
* carry forward events usually near but not adjacent to homopolymers | |||
'''GS20''' | |||
* 1.6M total wells | |||
* 450K detactable wells | |||
* 200K usable wells | |||
Accuracy: | |||
* published per-base accuracy of a Roche GS20 is only 96%. | |||
* Mitch Sogin paper | |||
* 99.5% accuracy rate in unassembled sequences | |||
* identified several factors that can be used to remove a small percentage of low-quality reads, improving the accuracy to 99.75% or better => better quality than Sanger sequencing | |||
* The error rate, defined as the number of errors (miscalled bases plus inserted and deleted bases) divided by the total number of expected bases, was 0.49% | |||
* 36% insertions, 27% delitions, 21% N's, 16% substitutions | |||
* A to G and T to C, were more frequent than other mismatches | |||
* reverse transitions, G to A and C to T, were not that frequent | |||
* Nearly 70% of the homopolymer extensions were A/T | |||
* errors were evenly distributed along the length of the reference sequences, they were not evenly distributed | |||
among reads: 82% had no errors, 93% had no more than a single error, and 96% had no more than 2 errors. | |||
* A small number of reads, fewer than 2%, contained a disproportionate number of errors that account for nearly 50% of the miscalls for the entire dataset | |||
* Avg quality is 25; in homopolymers can drop as low as 5 | |||
* Reads much longer than avg length had more errors | |||
* strong correlation between the presence of ambiguous base calls and other errors in a read | |||
* The presence of even a single ambiguous base in a read correlates strongly with the presence of other errors | |||
* Primer errors also correlated with errors | |||
'''GS FLX''' | |||
'''GS FLX with Titanium reagents''' | |||
* up to 500M/run | |||
* reads up to 500bp | |||
Get info from .sff files: | |||
$ sffinfo -h | |||
Usage: sffinfo [options...] [- | sfffile] [accno...] | |||
Options: | |||
-a or -accno Output just the accessions | |||
-s or -seq Output just the sequences | |||
-q or -qual Output just the quality scores | |||
-f or -flow Output just the flowgrams | |||
-t or -tab Output the seq/qual/flow as tab-delimited lines | |||
-n or -notrim Output the untrimmed sequence or quality scores | |||
-m or -mft Output the manifest text | |||
=== un-paired reads === | |||
=== paired ends === | |||
Features: | Features: | ||
| Line 9: | Line 119: | ||
* have a ~ 44-mer linker sequence in the middle | * have a ~ 44-mer linker sequence in the middle | ||
* flanked by a ~ 20-mer sequence on each side. | * flanked by a ~ 20-mer sequence on each side. | ||
* The two flanking 20-mers are segments of DNA that were originally located approximately 2.5 kb apart in the genome of interest. | * The two flanking 20-mers are segments of DNA that were originally located approximately 2.5 (3?) kb apart in the genome of interest. | ||
* The ordering and orienting of contigs generates scaffolds which provide a high-quality draft sequence of the genome. | * The ordering and orienting of contigs generates scaffolds which provide a high-quality draft sequence of the genome. | ||
Linker(palindrome) : GTTGGAACCGAAAGGGTTTGAATTCAAACCCTTTCGGTTCCAAC | |||
Check for linker : sffinfo -s *.sff | ~/bin/fasta2tab.pl | grep GTTGGAACCGAAAGGGTTTGAATTCAAACCCTTTCGGTTCCAAC | |||
12345678901234567890123456789012345678901234 | |||
GTTGGAACCGAAAGGGTTTGAATTCAAACCCTTTCGGTTCCAAC | |||
GTTGGAACCGA | |||
AAGGGTTTGAA | |||
TTCAAACCCTT | |||
TCGGTTCCAAC | |||
Anomalies: | Anomalies: | ||
* the linker can appear (tandem,completely/partially) more than once | * the linker can appear (tandem,completely/partially) more than once | ||
* some reads end up in linker (partial) | |||
* some reads don't contain the linker at all | |||
* some reads are cloning vector | |||
NCBI bacterial data sets: | |||
Accession Center Instrument | |||
SRR001351,2,3 JCVI 454 GS FLX 454 Paired End Sequencing Porphyromonas gingivalis W83 | |||
SRR001355 JCVI 454 GS FLX 454 Paired End Sequencing Escherichia coli str. K-12 MG1655 | |||
SRR004895,9 BI 454 GS FLX 45X on 454 (15X fragment and 30X paired) Brucella pinnipedialis | |||
SRR004900,1 BI 454 GS FLX 45X on 454 (15X fragment and 30X paired) Brucella suis bv. 3 | |||
SRR005309,10,11 BI 454 GS FLX 45X on 454 (15X fragment and 30X paired) Brucella ceti | |||
SRR005481,2 BI 454 GS FLX 45X on 454 (15X fragment and 30X paired) Brucella ceti BROAD:SEQUENCING_SAMPLE:24613.2 | |||
SRR005486,7,8 BI 454 GS FLX 45X on 454 (15X fragment and 30X paired) Brucella pinnipedialis BROAD:SEQUENCING_SAMPLE:246 | |||
SRR006465 GSC 454 GS FLX 454 Paired-End Library. Acinetobacter sp. ADP1 | |||
File location: | |||
/fs/szdata/454p/ | |||
/fs/szasmg2/Bacteria/Pseudomonas_syringae/Data/DC3000.format.454Reads.fna : Pseudomonas_syringae | |||
== Illumina == | |||
* Sequencing by Synthesis | |||
* Platforms: | |||
* Genome Analyzer (GA) | |||
* Genome Analyzer II : faster, higher tput | |||
* Future: 10GB/run 50bp reads | |||
* Future: 20GB/run 100bp reads | |||
Data sets: | |||
[ftp://ftp.sanger.ac.uk/pub/PRODUCTION_SOFTWARE/data_sets/suis_solexa/ Strep suis Solexa data set for download at Sanger] | [ftp://ftp.sanger.ac.uk/pub/PRODUCTION_SOFTWARE/data_sets/suis_solexa/ Strep suis Solexa data set for download at Sanger] | ||
[http://www.genomic.ch/edena/mw2Reads.seq.gz Staphylococcus aureus strain MW2 (edena paper)] | |||
[ftp://ftp.ncbi.nlm.nih.gov/pub/TraceDB/misc/examples/solexa NCBI Solexa example data set] | [ftp://ftp.ncbi.nlm.nih.gov/pub/TraceDB/misc/examples/solexa NCBI Solexa example data set] | ||
Pseudomonas aeruginosa | |||
[http://www. | Pseudomonas syringae | ||
NCBI SRA | |||
Applications: | |||
* Gene Expression | |||
* [http://www.biotechnews.com.au/index.php/id;13675486;fp;4;fpid;1017 ChIPSeq] (hight throughput) | |||
* Re-sequencing | |||
* mRNA sequencing | |||
Software: | Software: | ||
[http://sourceforge.net/projects/staden/ Staden & Io_lib] | |||
* IO_LIB package /fs/sz-user-supported/common/packages/io_lib-1.11-x86_64/bin/ | * IO_LIB package /fs/sz-user-supported/common/packages/io_lib-1.11-x86_64/bin/ | ||
* STADEN package /fs/sz-user-supported/common/packages/staden-src-1-7-0/distrib/unix-rel-1-7-0/linux-bin | * STADEN package /fs/sz-user-supported/common/packages/staden-src-1-7-0/distrib/unix-rel-1-7-0/linux-bin | ||
[http://maq.sourceforge.net/maq-man.shtml#download MAQ Sanger assembler] | |||
[http://www.bioperl.org/wiki/FASTQ_sequence_format FASTQ sequence format] | |||
Illumina 1G : | |||
* ~40 Million DNA sequencing reactions | |||
* about 36 hours for a run | |||
* each sequence is up to 36 bases long | |||
* insert len=~200bp | |||
Illumina Genome Analyzer II: | |||
* up to 51 bp | |||
* mate-pairs: opposite directions, slight overlap (insert size is less than 200bp "advertised") | |||
* on the SRA mate-pairs are joined; when downloaded only one read is shown. What about the mate pair? | |||
SRA: set of 4 files | |||
*_seq.txt : lane,run, well(x,y) sequence | |||
*_prb.txt : max quality from each group of 4 values is taken as quality | |||
*_sig2.txt : lane,run, well(x,y); max signal from each group of 4 values corresponds to max quality | |||
*_qhg.txt : lane,run, well(x,y); some encoded info? | |||
# *_seq.txt | |||
5 1 1269 1795 AAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAA | |||
# _prb.txt | |||
40 -40 -40 -40 40 -40 -40 -40 ... | |||
# _sig2.txt <== | |||
5 1 1269 1795 2594.0 2367.0 -10.0 -96.0 ... | |||
Qualities: | |||
Range : -5..40 | |||
Avg : ~25, depending on the data set | |||
=== Fastq format === | |||
[http://maq.sourceforge.net/fastq.shtml Maq help] | |||
Example: | Example: | ||
$ solexa2srf s_8_0100_seq.txt | 1 lane of Solexa reads: 10,959 READS; all are 36 bp | ||
$ srf2fastq | $ /fs/sz-user-supported/common/packages/io_lib-x86_64/bin/solexa2srf s_8_0100_seq.txt ; mv traces.srf s_8_0100.srf | ||
$ /fs/sz-user-supported/common/packages/io_lib-x86_64/bin/srf2fastq s_8_0100.srf > s_8_0100.fastq | |||
@s_8_100_293_551 | @s_8_100_293_551 | ||
| Line 47: | Line 234: | ||
+ | + | ||
IIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIII;>1 | IIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIII;>1 | ||
... | |||
+ | Edge effect: | ||
N's have quality -14 | |||
@ | |||
$ cat s_8_0100_seq.txt | sort -nk3 -nk4 | |||
+ | 8 100 0 37 ......AT.AT...TAATCAATA..GA.GAAG.... | ||
... | |||
8 100 1003 959 AGTC.......T.C.........GT.........AA | |||
$ more traces.qual | |||
... | |||
>s_8_100_0_37 | |||
-14 -14 -14 -14 -14 -14 25 13 -14 25 25 -14 -14 -14 25 25 25 25 22 25 25 25 25 -14 -14 25 25 -14 25 -11 25 14 -14 -14 -14 -14 | |||
... | |||
>s_8_100_1003_959 | |||
25 25 25 25 -14 -14 -14 -14 -14 -14 -14 25 -14 25 -14 -14 -14 -14 -14 -14 -14 -14 -14 25 -10 -14 -14 -14 -14 -14 -14 -14 -14 -14 8 25 | |||
... | |||
# bioperl script to convrt seq formats | |||
$ seqconvert.PLS --from fastq --to fasta < s_8_0100.fastq | |||
# get fastq qualities | |||
$ more *fastq | grep -A 1 "^+" | grep -v ^+ | grep -v -- ^-- | perl -ane '@F=split //,$F[0]; foreach (@F) { $n=ord($_)-33; print $n," ";} print "\n";' | |||
# convert Solexa format (maq fq_all2std.pl script) | |||
$ fq_all2std.pl seqprb2std s_5_0001_seq.txt s_5_0001_prb.txt > s_5_001.fastq | |||
$ fq_all2std.pl fq2fa s_5_001.fastq > s_5_001.seq | |||
# convert fastq to seq & qual | |||
fastq2seqqual.pl test.fastq | |||
fastq2seqqual.pl -solexa test.fastq | |||
=> test.seq , test.qual | |||
== SOLiD == | |||
* [http://www3.appliedbiosystems.com/AB_Home/applicationstechnologies/SOLiDSystemSequencing/index.htm ABI SOLiD] | |||
* [http://www.google.com/url?sa=t&source=web&ct=res&cd=1&url=http%3A%2F%2Fwww3.appliedbiosystems.com%2Fcms%2Fgroups%2Fmcb_marketing%2Fdocuments%2Fgeneraldocuments%2Fcms_057559.pdf&ei=gZg1Sc7SMoauevqTufYH&usg=AFQjCNHvV10RI-q4ORqN-2s0PXJ6GsBv1g&sig2=oK49We1BZ8naITjM3HxJ1A article] | |||
* [http://www3.appliedbiosystems.com/AB_Home/applicationstechnologies/SOLiDSystemSequencing/SoftwareCommunityDataAnalysisResourcesforScientistsDevelopers/index.htm Tools & Data Sets] | |||
* color space (0123) => base space (ACGT) | |||
* .csfsta file : in color space; start with a known base (usually T) | |||
Example: | |||
Ecoli_F3.csfasta | |||
>600_50_31_F3 | |||
T2222002113300322132112231 | |||
>600_50_63_F3 | |||
T2330133212130133221033110 | |||
Ecoli_F3_QV.qual | |||
>600_50_31_F3 | |||
15 20 14 11 25 20 17 21 16 9 15 12 21 8 2 10 15 5 3 5 10 6 2 4 4 | |||
>600_50_63_F3 | |||
5 13 9 23 5 9 8 4 6 4 5 4 7 6 10 7 7 5 13 11 5 8 2 7 6 | |||
Ecoli_R3.csfasta | |||
>600_51_85_R3 | |||
G0331332123123330101312331 | |||
>600_51_178_R3 | |||
G1111033111110111101111111 | |||
Ecoli_R3_QV.qual | |||
>600_51_85_R3 | |||
10 13 16 15 11 17 6 13 10 9 15 8 13 10 12 11 8 5 3 6 12 8 11 6 14 | |||
>600_51_178_R3 | |||
2 2 2 5 8 2 2 2 2 3 2 3 2 3 7 4 3 4 5 4 2 2 5 6 17 | |||
* low error rate (higher accuracy than Illumina) | |||
* 2008: 4G run, read_len=35bp; insert=3Kbp (old) | |||
* 2009: 9G run, read_len=50bp; | |||
* SOLiD™ 3 System generates (Oct 1 2008) | |||
** over 20 gigabases | |||
** mate-paired libraries with insert sizes ranging from 600 bp up to 10 kbp | |||
** human genome for less than $60,000. | |||
* uniform bases quality | |||
* accuracy greater than 99.94% | |||
* because of double base interogation & high cvg, qualities can be "discarded" | |||
Alignment matrix | |||
A C G T | |||
A 0 1 2 3 | |||
C 1 0 3 2 | |||
G 2 3 0 1 | |||
T 3 2 1 0 | |||
Examples: | |||
AA is encoded as 0 | |||
CG is encoded as 3 | |||
AACG is encoded as 0 1 3 | |||
Features of Color space: | |||
* Color space data are self-complementary | |||
Example: | |||
Base A G C T C G T C G T G C A G | |||
Color space 2 3 2 2 3 1 2 3 1 1 3 1 2 | |||
Complemented | |||
Base T C G A G C A G C A C G T C | |||
Color space 2 3 2 2 3 1 2 3 1 1 3 1 2 | |||
* Two-Base Encoding and Error Recognition | |||
1 change: measuring error | |||
multiple changes starting at a certain point: SNP | |||
Example: | |||
Reference 2 3 2 2 '''3''' 1 2 3 1 1 3 1 2 | |||
Observed 2 3 2 2 '''0''' 1 2 3 1 1 3 1 2 | |||
Data Sets: | |||
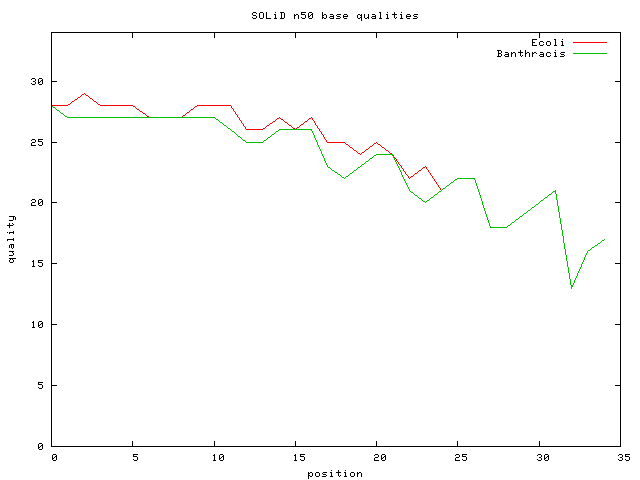
* [http://solidsoftwaretools.com/gf/project/dh10bmp/ E.Coli DH10B Mate-Pair Data Set] ~ 56M 25bp paired reads =~ 1G bp ; 56M*25/4.6M => 300X cvg | |||
* E.Coli DH10B Mate-Pair Data SubSet ~ 1.9M 25bp paired reads ; 10.17X cvg; avg inset is 2Kbp | |||
* [http://socs.biology.gatech.edu/Download.html Bacillus anthracis] 27M 35 bp unmated reads => 187X cvg | |||
[[Image:Pos-vs-quality.png]] | |||
== Helicos == | |||
* [http://www.helicosbio.com/ Web site] | |||
* [http://www.genomeweb.com/issues/news/151302-1.html @Broad] | |||
* 100Mbp/hour, 33bp reads, 5% error rate | |||
* [http://www.genomeweb.com/sequencing/helicos-slashes-instrument-price-25-percent-obtains-order-university-maryland UMD instrument] | |||
* [http://www.nature.com/nbt/journal/vaop/ncurrent/abs/nbt.1561.html Nature Biotech Aug 2009 article] | |||
* [http://www.ncbi.nlm.nih.gov/sites/entrez?db=sra&cmd=search&term=SRA009216 Human SRA data set] (not avail) | |||
* [http://www.ncbi.nlm.nih.gov/sra?term=helicos SRA data sets] (not avail) | |||
== Pacific Biosciences == | |||
* [http://www.pacificbiosciences.com/index.php Company website] | |||
* [http://www.bio-itworld.com/BioIT_Content.aspx?id=71746 BioIT article] | |||
* [http://www.sciencemag.org/cgi/content/abstract/1162986?ijkey=jDCYbOratwXCc&keytype=ref&siteid=sci Science paper] | |||
* [http://oelemento.wordpress.com/2011/01/03/a-closer-look-at-the-first-pacbio-sequence-dataset/ A closer look at the first PacBio sequence dataset BLOG] | |||
* [http://www.genomeweb.com/sequencing/pacbio-says-strobe-sequencing-increases-effective-read-length-single-molecule-se InSequenec article] May 2009 | |||
** strobe reads: 3kbp+ | |||
** Instead of generating a single, uninterrupted read of approximately 3.2 kilobases in length, by turning the lasers off and on during the run, one could double the size of the read, which consists of around 20 short "strobed" sequence reads that are interspersed with gaps. | |||
** Uninterrupted reads are limited to approximately 3,000 bases due to laser-induced photochemistry that damages the polymerases, he explained. However, when the lights are turned off, the polymerases keep incorporating nucleotides without incurring damage, and the available read length is "essentially put on hold, so you can take whatever read length you have and distribute it" over a longer stretch of DNA | |||
** Researchers could, for example, create the equivalent of a mate-pair read, with two sequences of more than a kilobase at each end and a long stretch of unidentified bases in between. Alternatively, they could cover a long piece of DNA with lots of short stitches of sequence. The length of these stitches and their distance can be varied without a need for different DNA libraries, Turner noted. | |||
* http://www.pacificbiosciences.com/newsletter_2011_05/informatics.php | |||
* http://www.pacbiodevnet.com/ login | |||
* http://www.pacbiodevnet.com/ConversionTools | |||
* [http://www.ncbi.nlm.nih.gov/sra/SRX032451?report=full SRX032451] SRA dataset; Sequencing for H1 (Haiti, 2010) | |||
* [http://www.pacbiodevnet.com/EColi_WF_DS Ecoli] Company dataset ; registration needed | |||
* http://www.genomeweb.com/informatics/pacbio-releases-open-source-analysis-suite-ahead-single-molecule-sequencer-launc | |||
* [http://www.pacbiodevnet.com/smrtanalysis/software/smrtanalysis smrtanalysis] | |||
=== Megachile rotundata PacBio sequence === | |||
** HDF5 (http://www.hdfgroup.org/HDF5/) This is PacBio's native data format, used to feed their primary and secondary pipelines. There is almost no support for this format outside of PacBio's analysis pipeline. Subsets of this data may be generated, however, as indicated below (e.g., FASTA and FASTQ format). According to the documentation (attached) for PacBio's hybrid assembly protocol (HybridAssembly) you may use HDF5 or FASTA in conjunction with Illumina-based scaffolds (or assembly-rpoduced contigs.) | |||
** High Quality (HQ) FASTA/FASTQ (post-primary analysis) We can derive from the HDF5 file FASTA and FASTQ sequences for reads passing primary analysis in the PacBio pipeline. These reads have essentially been "okayed" by the pipeline as high quality reads to carry forward in secondary analysis--such as attempted alignment to a reference genome, etc. These reads have not had PacBio's proprietary SMRT bell adaptor (45 nt x 2) trimmed... as trimming is part of the downstream PacBio secondary analysis pipeline. These are single pass representations of the sequence reads and therefore contain uncorrected errors, insertions, and deletions incidental to the current state of the PacBio technology. The FASTQ files have PacBio's "Phred-like" quality values corresponding to the nucleotide representation of the sequence. In the instances where a SMRT bell molecule is sequenced multiple times (i.e., multiple passes) each pass is termed a "sub-read". In this representation of the data, sub-reads have not been broken out from the parent sequence. | |||
** High Quality (HQ) FASTA (edited) These FASTA files are similar to the ones described above, but have been filtered and edited. Here, the SMRT bell adaptors have been excised from the parent reads, and entries have been broken into corresponding sub-reads. Only reads with a quality filter of 75% or better have been retained; sub-reads less than 50 nt in length have been removed. These are still single pass representations of the sequence reads and therefore contain uncorrected errors, insertions, and deletions incidental to the current state of the PacBio technology. | |||
** [[Media:SMRT_Pipe_Reference_Guide.pdf]] | |||
== Visigen == | |||
== Polonator == | |||
* [http://www.polonator.org/index.htm Web Site] | |||
== RainDance Technologies == | |||
* [http://www.raindancetechnologies.com/ Web site] | |||
== Whole genome profiling (Wgp) == | |||
* [http://www.keygene.com/services/technologies_whole_genome_profiling.php keygene] | |||
''In our view getting to a good quality assembly especially in large genomes is a real challenge and therefore I would like to get your attention to our Whole Genome Profiling technology that generated a lot of interest at the last PAG meeting in San Diego, where we presented the technology in several workshops. | |||
WGP has now been used to help the assembly of many of the plant genomes for Melon, Tomato, Potato, Lettuce, Brassica napus, Brassica rapa, Brassica oleraceae, Strawberry, Tobacco, Wheat, and Sunflower. | |||
Also at the moment we are working on a number of genomes both for academic labs/consortia as well as for industry. | |||
The power of the WGP technology lies in the creation of a sequence-based scaffold of the genome based on the creating of sequence tags in BAC libraries (average 30 tags pet BAC). | |||
This provides ordered BAC clones covering the genome on which shot-gun sequence information can be placed. | |||
WGP solves the problem of genome assembly that most laboratories are struggling with because it simplifies the bio-informatics effort required to assemble shot-gun sequence information considerably. | |||
For instance, shot-gun sequencing of the melon genome (450 Mb) left us with 13,486 contigs, while the WGP map for Melon gave us BAC-based 1085 contigs. | |||
By putting these two methods together, we ended up with a very high quality reference amp of only 249 supercontigs. | |||
In addition, the BAC clones linked to the physical map allows you to go directly to clones of those regions of the genome you are interested in. | |||
I have included three relevant abstracts of PAG presentations in the attachment for your reference. | |||
In short: | |||
The assembly of the tetraploid tobacco genome (4.5 Gb) was presented at the PAG and worked perfectly well, showing that genome size does not influence the quality of the WGP maps (see abstract below) and in addition showed | |||
that the WGP assemblies are very accurate as concluded from the fact that 97% of the WGP contigs can be assigned to one of the ancestral genomes that are part of the tetraploid tobacco genome. | |||
Also the progress on the Sunflower genome (3.6 Gb) was presented and my colleague Michiel van Eijk showed the power of the technology to deal with repetitivegenomes as well a new approach on genome sequence assembly that takes as a starting oint a good quality WGP physical map. | |||
Taking all this together and the fact that the technical paper on WGP was just accepted forpublication in Genome Research (preprint included in attachment), I think this technology may be of interest for you to improve your genome assembly as well.'' | |||
== Ion Torrent == | |||
* [http://www.ncbi.nlm.nih.gov/sra/SRX067313?report=full SRA Escherichia coli O104:H4 str. TY-2482] | |||
* [http://ioncommunity.iontorrent.com/docs/DOC-1516 Company Escherichia coli O104:H4 str. TY-2482] | |||
* SImilar to 454; indel & homopolimer issues | |||
= Download = | |||
== Genbank == | |||
* Bioperl scripts: | |||
bp_fetch.pl net::genbank:NC_005810.1 > NC_005810.1 | |||
bp_fetch.pl net::genbank:NC_005810 > NC_005810 | |||
bp_fetch.pl net::genbank:45439865 > 45439865 | |||
* NCBI utilities: | |||
wget "http://eutils.ncbi.nlm.nih.gov/entrez/eutils/epost.fcgi?dbfrom=nucleotide&id=257481614" -O - | grep WebEnv | p 'next unless /<WebEnv>(.+)<\/WebEnv>/; echo "setenv WebEnv $1\n";' | |||
wget "http://eutils.ncbi.nlm.nih.gov/entrez/eutils/efetch.fcgi?db=nucleotide&query_key=1&WebEnv=NCID_1_35952872_130.14.18.46_9001_1260305427" -O - | |||
== TA == | |||
* [http://www.ncbi.nlm.nih.gov/Traces/trace.cgi?cmd=stat&f=xml_list_arrivals_retrievals&m=main&s=main TA] | |||
* [ftp://ftp.ncbi.nih.gov/pub/TraceDB/ TA FTP] | |||
* Example: | |||
query_tracedb "query text species_code='BACILLUS ANTHRACIS'" > ids.bacillus_anthracis.001 | |||
( echo -n "retrieve_tgz fasta quality xml " ; cat ids.bacillus_anthracis.001 | head -40000 | paste -s -d "," ) \ | |||
| query_tracedb > bacillus_anthracis.001.1.tgz | |||
( echo -n "retrieve_tgz fasta quality xml " ; cat ids.bacillus_anthracis.001 | head -80000 | tail -40000 | paste -s -d "," ) \ | |||
| query_tracedb > bacillus_anthracis.001.2.tgz | |||
... | |||
== SRA == | |||
* [http://www.ncbi.nlm.nih.gov/Traces/sra/sra.cgi? SRA] | |||
= Format = | |||
== FASTA == | |||
Utilities: | |||
* Indexing large files | |||
#NCBI utilies: expect a certain id format Ex: ">lcl|12345" | |||
formatdb -i prefix.fasta -p F -o T | |||
fastacmd -d prefix.fasta -p F -s id1,id2,... | |||
fastacmd -d prefix.fasta -p F -i prefix.filter | |||
~/bin/formatdb.pl < prefix.fasta > prefix.fasta.offset | |||
~/bin/fastacmd.pl -s 1,2 -o prefix.fasta.offset < prefix.fasta > subset.fasta | |||
== TA == | |||
* Sanger: | |||
tarchive2amos -o Ba Ba.seq # TA FTP | |||
tarchive2amos -o Ba -tracedir traces/ # TA querytrace_db | |||
tarchive2amos -o Ba -assembly assembly/ASSEMBLY.xml -tracedir traces/ # AA | |||
== SRA == | |||
* Solexa mated: | |||
seq2amos.pl -n solexap -m 100 -s 20 -fs solexa_f.fasta -rs solexa_f.fasta > solexap.afg | |||
seq2amos.pl -n solexap -m 100 -s 20 -fs solexa_f.fasta -fq solexa_f.qual -rs solexa_r.fasta -rq solexa_r.qual > solexap.afg | |||
seq2amos.pl -n solexap -m 100 -s 20 -fs solexa_f*.fasta -rs solexa_r*.fasta > solexap.afg | |||
* Solexa unmated: | |||
seq2amos.pl -n solexa -fs solexa.fasta > solexa.afg | |||
* 454 mated: | |||
seq2amos.pl -n 454p -m 3000 -s 300 -fs 454.seq > 454p.afg | |||
* 454 unmated: | |||
seq2amos.pl -n 454 -fs 454.seq > 454.afg | |||
= Convestion = | |||
* Readseq | |||
~/bin//readseq.sh -f Fasta -o prefix.fasta prefix.embl | |||
* Bioperl | |||
bp_sreformat.pl -i prefix.embl -o prefix.fasta -if EMBL -of Fasta | |||
* AMOS: | |||
amos2frg [-i infile] [-o outfile] | |||
amos2sq [-i] infile [-o outprefix] => outprefix.seq outprefix.qual | |||
= Read Simulators = | |||
* Metasim | |||
= Read Mapping Software = | |||
* [http://en.wikipedia.org/wiki/Sequence_alignment_software#Short-Read_Sequence_Alignment Short-Read_Sequence_Alignment programs] | |||
== BFAST == | |||
* need to e-mail to author to get the code | |||
== BLAT == | |||
* [http://www.genome.org/cgi/reprint/GR-2292Rv1 BLAT—The BLAST-Like Alignment Tool, Genome Research 2002] | |||
* [http://genome.ucsc.edu/FAQ/FAQblat FAQ] | |||
* Can align any type of reads | |||
* Can do nt:aa translation | |||
* Command: blat | |||
blat -noHead -t=dna -q=dna -tileSize=10 -stepSize=3 Pa.1con Pa.seq Pa.blat | |||
== BLASTALL == | |||
* Env vars | |||
echo $BLASTMAT $BLOSUM62 $BLASTDB | |||
/fs/sz-user-supported/Linux-x86_64/packages/blast-2.2.18/data/ | |||
/fs/sz-user-supported/Linux-x86_64/packages/blast-2.2.18/data/BLOSUM62 | |||
/fs/szdata/ncbi/ftp.ncbi.nih.gov/blast/db/ | |||
* Example | |||
blastall -p blastn -d refseq_genomic -i acc.keep.fna | |||
== BOWTIE == | |||
* build index: | |||
bowtie-build prefix.1con prefix | |||
=> | |||
prefix.rev.2.ebwt | |||
prefix.rev.1.ebwt | |||
prefix.2.ebwt | |||
prefix.1.ebwt | |||
prefix.3.ebwt | |||
prefix.4.ebwt | |||
* run alignments | |||
bowtie prefix qry.fastq -a --best --strata -n 2 -l 28 -e 70 | |||
== MAQ/BWA == | |||
* [http://bio-bwa.sourceforge.net/bwa.shtml BWA] | |||
== RMAP == | |||
* [http://rulai.cshl.edu/rmap/ RMAP] : designed for Illumina-Solexa | |||
* Command: rmap | |||
rmap -m 3 -w 33 -c Pa.1con Pa.seq -o Pa.rmap | |||
== SHRiMP == | |||
* [http://compbio.cs.toronto.edu/shrimp/ Web site] | |||
* Commands: rmapper-cs , rmapper-ls, ... | |||
== SeqMap == | |||
* [http://biogibbs.stanford.edu/~jiangh/SeqMap/ SeqMap] developed at Stanford | |||
* allows up to five mixed substitutions and inserted/deleted nucleotides in the mapping | |||
* allows sequences to contain N’s, and to have unequal lengths | |||
./seqmap | |||
Usage: seqmap <number of mismatches> <probe FASTA file name> <transcript FASTA file name> <output file name> [options] | |||
Parameters: | |||
<number of mismatches> maximum edit distance allowed | |||
<probe FASTA file name> probe/tag/read sequences | |||
<transcript FASTA file name> reference sequences | |||
<output file name> name of the output file | |||
... | |||
== SHORE == | |||
* [http://1001genomes.org/downloads/ SHORE] | |||
== SOAP == | |||
* [http://soap.genomics.org.cn/ Web site (China)] | |||
* [http://soap.genomics.org.cn/#Formatofoutput Formatofoutput] | |||
* [http://soap.genomics.org.cn/SOAP_paper.pdf SOAP: short oligonucleotide alignment program, Bioinformatics Jan 2008] | |||
* Commands: soap, soap.contig, soap_dealign, soap.huge, soap.short | |||
* can use qualities, do read trimming, use pair ends, RNA alignments | |||
soap -v <max_mismatches> -g <max_gap> -r <repeat_behavior> -p <processors> -d ref.fasta -a qry.fasta -o ref-qry.soap | |||
== SOAPaligner/soap2 == | |||
* ~10 times faster than SOAP | |||
/nfshomes/dpuiu/szdevel/soap2.20release/2bwt-builder ref.fasta # => ref.fasta.index | |||
soap2 -D ref.fasta.index -a qry.fasta -o qry.soap -r <repeat_behavior> -l <seed_len> -p <processors> | |||
soap2 -D /scratch1/Bombus_impatiens/Data/mito.1con.index -a s_3_1_sequence.seq -o s_3_1_sequence.soap2 -r 2 -l 36 -p 16 # up to 2 mismatches in the first 36bp | |||
== SOCS == | |||
* [http://socs.biology.gatech.edu/ Web site] | |||
* ABI color space | |||
socs socs.pref | |||
more socs.pref | |||
Req.fa | |||
Seq_F3.csfasta | |||
Seq_F3_QV.qual | |||
out_prefix | |||
2 | |||
1000 | |||
2 | |||
false | |||
true | |||
0 | |||
== SOLiD == | |||
* [http://solidsoftwaretools.com/gf/project/corona/ SOLID System Analysis Pipeline Tool (Corona Lite)] | |||
== SSAHA == | |||
* [http://www.sanger.ac.uk/Software/analysis/SSAHA/ Web site(Sanger)] | |||
* Focused on exact, nearly exact matches | |||
* Does not find all the exact matches??? | |||
* Example: Solexa 33bp ~30% of reads are not found | |||
== ZOOM == | |||
* [http://bioinformatics.oxfordjournals.org/cgi/content/full/24/21/2431 ZOOM] | |||
== Whole genomes alignments == | |||
* BLAT | |||
* BLASTZ, [http://www.ncbi.nlm.nih.gov/pubmed/10745991 Post-processing long pairwise alignments article.] decom program | |||
* LAGAN | |||
* MUMMER | |||
* AVID | |||
= Genome Resequencing = | |||
* [http://www.nature.com/nature/journal/v452/n7189/pdf/nature06884.pdf The complete genome of an individual by massively parallel DNA sequencing (J.Watson's genome) Nature April 2008] | |||
* [http://www.nature.com/nature/journal/v452/n7189/extref/nature06884-s1.pdf J.Watson's genome (supplementary info) ] | |||
* [http://www.nature.com/nmeth/journal/v5/n2/full/nmeth.1179.html;jsessionid=DC518BCD8B2CACAE8AFFF7F70DD46902 Whole-genome sequencing and variant discovery in C. elegans Nature Jan 2008] | |||
= Data sets = | |||
== Solexa == | |||
* [ftp://ftp.sanger.ac.uk/pub/PRODUCTION_SOFTWARE/data_sets/suis_solexa/ Strep suis Solexa at Sanger] 2,659,250 36bp single reads, ~49X coverage | |||
/fs/szdata/Solexa/Streptococcus_suis/suisp17/strip3.fna | |||
Velvet (0.7.55) assembly: | |||
. elem min q1 q2 q3 max mean n50 sum misassembled | |||
seq 2659250 36 36 36 36 36 36 36 95733000 . | |||
ctg 669 45 69 383 4314 32213 2944 8437 1969901 8 | |||
time 153.166u 4.673s 2:20.85 112.0% 0+0k 0+0io 2pf+0w | |||
SOAPdenovo (1.03) assembly: | |||
. elem min q1 q2 q3 max mean n50 sum misassembled | |||
seq 2659250 36 36 36 36 36 36 36 95733000 | |||
ctg 2219 24 25 32 93 24421 907 7209 2014315 1 | |||
ctg45 866 45 65 214 3307 24421 2282 7275 1976767 1 | |||
time 89.614u 1.599s 1:36.51 94.4% 0+0k 0+0io 0pf+0w | |||
SOAPdenovo vs Velvet: | |||
* slightly shorted ctgs | |||
* fewer errors (1 vs 8) | |||
* shorter run time (~half) | |||
* velvet example: 142,858 35bp reads that should assemble into a ~100Kbp contig | |||
/nfshomes/dpuiu/szdevel/velvet/data/test_reads.fa | |||
* [http://www.genomic.ch/edena/mw2Reads.seq.gz Staphylococcus aureus strain MW2 (edena paper)] 35bp, ~47X coverage | |||
* Pseudomonas aeruginosa: 33bp, ~43X coverage | |||
* Pseudomonas syringae: 32bp, ~31X coverage | |||
* 1000 Genomes (June 14th 2008): 47bp | |||
Accession #Runs Instrument Center Study [Individual] | |||
SRA000303 41 Solexa 1G Genome Analyzer BI 1000Genomes Project Pilot 2 NA12878 | |||
SRA000304 49 Solexa 1G Genome Analyzer BI 1000Genomes Project Pilot 2 NA12891 | |||
SRA000305 56 Solexa 1G Genome Analyzer BI 1000Genomes Project Pilot 2 NA12892 | |||
SRA000307 1 Solexa 1G Genome Analyzer SC 1000Genomes Project Pilot 1 NA10851 | |||
SRA000308 2 Solexa 1G Genome Analyzer SC 1000Genomes Project Pilot 1 NA11993 | |||
SRA000309 3 Solexa 1G Genome Analyzer SC 1000Genomes Project Pilot 1 NA11995 | |||
SRA000310 1 Solexa 1G Genome Analyzer SC 1000Genomes Project Pilot 1 NA12006 | |||
SRA000311 1 Solexa 1G Genome Analyzer SC 1000Genomes Project Pilot 1 NA12044 | |||
SRA000312 2 Solexa 1G Genome Analyzer SC 1000Genomes Project Pilot 1 NA12156 | |||
SRA000313 1 Solexa 1G Genome Analyzer SC 1000Genomes Project Pilot 1 NA12414 | |||
SRA000314 1 Solexa 1G Genome Analyzer SC 1000Genomes Project Pilot 1 NA12776 | |||
SRA000315 1 Solexa 1G Genome Analyzer SC 1000Genomes Project Pilot 1 NA12828 | |||
SRA000316 12 Solexa 1G Genome Analyzer SC 1000Genomes Project Pilot 2 NA12878 | |||
SRA000317 8 Solexa 1G Genome Analyzer SC 1000Genomes Project Pilot 2 NA12891 | |||
SRA000318 14 Solexa 1G Genome Analyzer SC 1000Genomes Project Pilot 2 NA12892 | |||
SRA000319 1 Solexa 1G Genome Analyzer SC 1000Genomes Project Pilot 1 NA12004 | |||
June 14th 2008: Sept 19th 2008 | |||
SRA001100 23 Illumina Genome Analyzer BGI 1000Genomes Project Pilot 2 NA19240 | |||
... | |||
SRA002029 1 Illumina Genome Analyzer II WUGSC 1000Genomes Project Pilot 2 NA19239 | |||
/fs/szdata/Solexa/1000genomes | |||
* Example SRR001113.seq : | |||
7,058,926 47 bp sequences | |||
2,402,398 contain at least 1 '.' | |||
== 454 == | |||
* 1000 Genomes | |||
June 14th 2008 | |||
Accession #Runs Instrument Center Study [Individual] | |||
SRA000302 121 454 GS FLX BCM 1000Genomes Project Pilot 2 NA12878 | |||
SRA001032 2 454 GS FLX BCM 1000Genomes Project Pilot 2 NA12878 | |||
SRA001036 1 454 GS FLX BCM 1000Genomes Project Pilot 1 NA12812 | |||
SRA001094 1 454 GS FLX BCM 1000Genomes Project Pilot 2 NA12878 | |||
June 14th 2008: Sept 19th 2008 | |||
SRA001037 2 454 GS FLX BCM 1000Genomes Project Pilot 1 NA12812 | |||
... | |||
SRA001819 1 454 GS FLX BCM 1000Genomes Project Pilot 2 NA12878 | |||
* Cryptosporidium_muris_RN66: SRA001029 (not paired) | |||
* EcoliK12: SRR001355 (paired) | |||
* Porphyromonas_gingivalis_W83: E8YURXS01 (paired) | |||
== Refseq == | |||
* /fs/szdata/genomes/human_ncbi_build36/ NCBI build36.1 May 2006 (Current build is 36.3 March 2008) | |||
* /fs/szdata/genomes/human_celera_2001_Orig/ | |||
= Links = | |||
* [http://www.1000genomes.org/page.php?page=home 1000 genomes] | |||
* [http://www.cbcb.umd.edu/~langmead/solexa_1000genomes.html Ben's web site 1] | |||
* [http://www.cbcb.umd.edu/~langmead/solexa_format.html Ben's web site 2] | |||
* [http://en.wikipedia.org/wiki/Chip-Sequencing Chip-Seq @ Wikipedia] | |||
* [http://www.ncbi.nlm.nih.gov/Traces/sra/sra.cgi?cmd=table&f=run&m=data&s=run SRA] | |||
* [ftp://ftp.ncbi.nih.gov/pub/TraceDB/ShortRead SRA FTP] | |||
Latest revision as of 14:19, 20 July 2011
Articles
- The impact of next-generation sequencing technology on genetics Elaine Mardis Trends in Genetics 2008
- As Users Demand Paired-End Sequencing, 454, Illumina, and ABI Work On New Kits
- Sequence Format Descriptions(EMBL)
- ismb2007Poster.pdf
- Smith_Rennes_2007.pdf
- GenomeAnalyzer_SpecSheet.pdf
- Assembly and Alignment Algorithms for Next-Gen Sequence Data
- Sequencing of natural strains of Arabidopsis thaliana with short reads (Illumina)
- Accuracy and quality of massively parallel DNA pyrosequencing
- Advanced sequencing technologies and their wider impact in microbiology
- Emerging technologies in DNA sequencing
- Lander Waterman
- Whole-genome re-sequencing
- Whole-Genome Sequencing and Assembly with High-Throughput, Short-Read Technologies
- seqanswers
- politigenomics table
- Valex & SRA
- BIOINFORMATICS FOR NEXT GENERATION SEQUENCING
- The Short Read Headache
- The impact of next-generation sequencing technology on genetics
- Next-generation DNA sequencing methods.
- Short-Read Sequencing Technologies for Transcriptional Analyses.
- Next Generation Sequencing - Bioinformatics Journal - Virtual Edition
- Sanger Who Science April 2009
- Accurate and comprehensive sequencing of personal genomes Genome Research July 2011
Cleaning
- run dust on Ecoli & UniVector to mask low complexity seqs
- screen against Ecoli & UniVector database
Technologies
Latest Technology Summary
Technology 454 Illumina Solid
seq-by-synthesis seq-by-synthesis ABI Company 454(Roche) Illumina Location Brandford,CT SanDiego,CA Latest GS FLX, Titanium reagents Genome Analyzer II SOLID 3
Throughput 500M/run 1G/run 20G/run RunTime 10hr 3days ReadLen 500bp 36 35 InsertLen 3K 100-200bp 600-10K
Accuracy 99.94%
Q20(99%accuracy) 400bp 34bp
Cost $3K/run 60K/3G
$400/4M bacterial genome(25-30X)
Problems homopolimers
Sanger
454
- Pyrosequencing
- 454_Life_Sciences wikipedia
- In-sequenece article
- 1_paired_end.pdf
- [1]
- [2]
Anomalies:
* homopolymer lengths can be shorter than real * substitutions less likely than in traditional methodssingle base insertions * carry forward events usually near but not adjacent to homopolymers
GS20
* 1.6M total wells * 450K detactable wells * 200K usable wells
Accuracy: * published per-base accuracy of a Roche GS20 is only 96%. * Mitch Sogin paper * 99.5% accuracy rate in unassembled sequences * identified several factors that can be used to remove a small percentage of low-quality reads, improving the accuracy to 99.75% or better => better quality than Sanger sequencing * The error rate, defined as the number of errors (miscalled bases plus inserted and deleted bases) divided by the total number of expected bases, was 0.49% * 36% insertions, 27% delitions, 21% N's, 16% substitutions * A to G and T to C, were more frequent than other mismatches * reverse transitions, G to A and C to T, were not that frequent * Nearly 70% of the homopolymer extensions were A/T * errors were evenly distributed along the length of the reference sequences, they were not evenly distributed
among reads: 82% had no errors, 93% had no more than a single error, and 96% had no more than 2 errors.
* A small number of reads, fewer than 2%, contained a disproportionate number of errors that account for nearly 50% of the miscalls for the entire dataset * Avg quality is 25; in homopolymers can drop as low as 5 * Reads much longer than avg length had more errors * strong correlation between the presence of ambiguous base calls and other errors in a read * The presence of even a single ambiguous base in a read correlates strongly with the presence of other errors * Primer errors also correlated with errors
GS FLX
GS FLX with Titanium reagents
* up to 500M/run * reads up to 500bp
Get info from .sff files:
$ sffinfo -h
Usage: sffinfo [options...] [- | sfffile] [accno...]
Options:
-a or -accno Output just the accessions
-s or -seq Output just the sequences
-q or -qual Output just the quality scores
-f or -flow Output just the flowgrams
-t or -tab Output the seq/qual/flow as tab-delimited lines
-n or -notrim Output the untrimmed sequence or quality scores
-m or -mft Output the manifest text
un-paired reads
paired ends
Features:
* approximately 84-nucleotide DNA fragments * have a ~ 44-mer linker sequence in the middle * flanked by a ~ 20-mer sequence on each side. * The two flanking 20-mers are segments of DNA that were originally located approximately 2.5 (3?) kb apart in the genome of interest. * The ordering and orienting of contigs generates scaffolds which provide a high-quality draft sequence of the genome.
Linker(palindrome) : GTTGGAACCGAAAGGGTTTGAATTCAAACCCTTTCGGTTCCAAC Check for linker : sffinfo -s *.sff | ~/bin/fasta2tab.pl | grep GTTGGAACCGAAAGGGTTTGAATTCAAACCCTTTCGGTTCCAAC 12345678901234567890123456789012345678901234 GTTGGAACCGAAAGGGTTTGAATTCAAACCCTTTCGGTTCCAAC
GTTGGAACCGA AAGGGTTTGAA TTCAAACCCTT TCGGTTCCAAC
Anomalies:
* the linker can appear (tandem,completely/partially) more than once * some reads end up in linker (partial) * some reads don't contain the linker at all * some reads are cloning vector
NCBI bacterial data sets:
Accession Center Instrument SRR001351,2,3 JCVI 454 GS FLX 454 Paired End Sequencing Porphyromonas gingivalis W83 SRR001355 JCVI 454 GS FLX 454 Paired End Sequencing Escherichia coli str. K-12 MG1655 SRR004895,9 BI 454 GS FLX 45X on 454 (15X fragment and 30X paired) Brucella pinnipedialis SRR004900,1 BI 454 GS FLX 45X on 454 (15X fragment and 30X paired) Brucella suis bv. 3 SRR005309,10,11 BI 454 GS FLX 45X on 454 (15X fragment and 30X paired) Brucella ceti SRR005481,2 BI 454 GS FLX 45X on 454 (15X fragment and 30X paired) Brucella ceti BROAD:SEQUENCING_SAMPLE:24613.2 SRR005486,7,8 BI 454 GS FLX 45X on 454 (15X fragment and 30X paired) Brucella pinnipedialis BROAD:SEQUENCING_SAMPLE:246 SRR006465 GSC 454 GS FLX 454 Paired-End Library. Acinetobacter sp. ADP1
File location:
/fs/szdata/454p/ /fs/szasmg2/Bacteria/Pseudomonas_syringae/Data/DC3000.format.454Reads.fna : Pseudomonas_syringae
Illumina
- Sequencing by Synthesis
- Platforms:
* Genome Analyzer (GA) * Genome Analyzer II : faster, higher tput * Future: 10GB/run 50bp reads * Future: 20GB/run 100bp reads
Data sets:
Strep suis Solexa data set for download at Sanger Staphylococcus aureus strain MW2 (edena paper) NCBI Solexa example data set Pseudomonas aeruginosa Pseudomonas syringae NCBI SRA
Applications:
* Gene Expression * ChIPSeq (hight throughput) * Re-sequencing * mRNA sequencing
Software:
Staden & Io_lib * IO_LIB package /fs/sz-user-supported/common/packages/io_lib-1.11-x86_64/bin/ * STADEN package /fs/sz-user-supported/common/packages/staden-src-1-7-0/distrib/unix-rel-1-7-0/linux-bin MAQ Sanger assembler FASTQ sequence format
Illumina 1G :
* ~40 Million DNA sequencing reactions * about 36 hours for a run * each sequence is up to 36 bases long * insert len=~200bp
Illumina Genome Analyzer II:
* up to 51 bp * mate-pairs: opposite directions, slight overlap (insert size is less than 200bp "advertised") * on the SRA mate-pairs are joined; when downloaded only one read is shown. What about the mate pair?
SRA: set of 4 files
*_seq.txt : lane,run, well(x,y) sequence *_prb.txt : max quality from each group of 4 values is taken as quality *_sig2.txt : lane,run, well(x,y); max signal from each group of 4 values corresponds to max quality *_qhg.txt : lane,run, well(x,y); some encoded info?
# *_seq.txt 5 1 1269 1795 AAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAA # _prb.txt 40 -40 -40 -40 40 -40 -40 -40 ... # _sig2.txt <== 5 1 1269 1795 2594.0 2367.0 -10.0 -96.0 ...
Qualities:
Range : -5..40 Avg : ~25, depending on the data set
Fastq format
Example:
1 lane of Solexa reads: 10,959 READS; all are 36 bp $ /fs/sz-user-supported/common/packages/io_lib-x86_64/bin/solexa2srf s_8_0100_seq.txt ; mv traces.srf s_8_0100.srf $ /fs/sz-user-supported/common/packages/io_lib-x86_64/bin/srf2fastq s_8_0100.srf > s_8_0100.fastq @s_8_100_293_551 CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCACC + IIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIII @s_8_100_35_698 TATATGATTGACAATATAAAAATATGAGTATAAAAT + IIIIIIIIIIIIIIIIIIIIIIIIIIIIIIII4/:I @s_8_100_880_947 TTATTATCTTTATTGACGTACCTCTAGAAGACCCAA + IIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIII;>1 ...
Edge effect: N's have quality -14
$ cat s_8_0100_seq.txt | sort -nk3 -nk4 8 100 0 37 ......AT.AT...TAATCAATA..GA.GAAG.... ... 8 100 1003 959 AGTC.......T.C.........GT.........AA
$ more traces.qual ... >s_8_100_0_37 -14 -14 -14 -14 -14 -14 25 13 -14 25 25 -14 -14 -14 25 25 25 25 22 25 25 25 25 -14 -14 25 25 -14 25 -11 25 14 -14 -14 -14 -14 ... >s_8_100_1003_959 25 25 25 25 -14 -14 -14 -14 -14 -14 -14 25 -14 25 -14 -14 -14 -14 -14 -14 -14 -14 -14 25 -10 -14 -14 -14 -14 -14 -14 -14 -14 -14 8 25 ...
# bioperl script to convrt seq formats
$ seqconvert.PLS --from fastq --to fasta < s_8_0100.fastq
# get fastq qualities
$ more *fastq | grep -A 1 "^+" | grep -v ^+ | grep -v -- ^-- | perl -ane '@F=split //,$F[0]; foreach (@F) { $n=ord($_)-33; print $n," ";} print "\n";'
# convert Solexa format (maq fq_all2std.pl script) $ fq_all2std.pl seqprb2std s_5_0001_seq.txt s_5_0001_prb.txt > s_5_001.fastq $ fq_all2std.pl fq2fa s_5_001.fastq > s_5_001.seq
# convert fastq to seq & qual fastq2seqqual.pl test.fastq fastq2seqqual.pl -solexa test.fastq => test.seq , test.qual
SOLiD
- ABI SOLiD
- article
- Tools & Data Sets
- color space (0123) => base space (ACGT)
- .csfsta file : in color space; start with a known base (usually T)
Example:
Ecoli_F3.csfasta >600_50_31_F3 T2222002113300322132112231 >600_50_63_F3 T2330133212130133221033110
Ecoli_F3_QV.qual >600_50_31_F3 15 20 14 11 25 20 17 21 16 9 15 12 21 8 2 10 15 5 3 5 10 6 2 4 4 >600_50_63_F3 5 13 9 23 5 9 8 4 6 4 5 4 7 6 10 7 7 5 13 11 5 8 2 7 6
Ecoli_R3.csfasta >600_51_85_R3 G0331332123123330101312331 >600_51_178_R3 G1111033111110111101111111
Ecoli_R3_QV.qual >600_51_85_R3 10 13 16 15 11 17 6 13 10 9 15 8 13 10 12 11 8 5 3 6 12 8 11 6 14 >600_51_178_R3 2 2 2 5 8 2 2 2 2 3 2 3 2 3 7 4 3 4 5 4 2 2 5 6 17
- low error rate (higher accuracy than Illumina)
- 2008: 4G run, read_len=35bp; insert=3Kbp (old)
- 2009: 9G run, read_len=50bp;
- SOLiD™ 3 System generates (Oct 1 2008)
- over 20 gigabases
- mate-paired libraries with insert sizes ranging from 600 bp up to 10 kbp
- human genome for less than $60,000.
- uniform bases quality
- accuracy greater than 99.94%
- because of double base interogation & high cvg, qualities can be "discarded"
Alignment matrix
A C G T A 0 1 2 3 C 1 0 3 2 G 2 3 0 1 T 3 2 1 0
Examples:
AA is encoded as 0 CG is encoded as 3 AACG is encoded as 0 1 3
Features of Color space:
* Color space data are self-complementary
Example:
Base A G C T C G T C G T G C A G
Color space 2 3 2 2 3 1 2 3 1 1 3 1 2
Complemented
Base T C G A G C A G C A C G T C
Color space 2 3 2 2 3 1 2 3 1 1 3 1 2
* Two-Base Encoding and Error Recognition 1 change: measuring error multiple changes starting at a certain point: SNP
Example:
Reference 2 3 2 2 3 1 2 3 1 1 3 1 2
Observed 2 3 2 2 0 1 2 3 1 1 3 1 2
Data Sets:
- E.Coli DH10B Mate-Pair Data Set ~ 56M 25bp paired reads =~ 1G bp ; 56M*25/4.6M => 300X cvg
- E.Coli DH10B Mate-Pair Data SubSet ~ 1.9M 25bp paired reads ; 10.17X cvg; avg inset is 2Kbp
- Bacillus anthracis 27M 35 bp unmated reads => 187X cvg
Helicos
- Web site
- @Broad
- 100Mbp/hour, 33bp reads, 5% error rate
- UMD instrument
- Nature Biotech Aug 2009 article
- Human SRA data set (not avail)
- SRA data sets (not avail)
Pacific Biosciences
- Company website
- BioIT article
- Science paper
- A closer look at the first PacBio sequence dataset BLOG
- InSequenec article May 2009
- strobe reads: 3kbp+
- Instead of generating a single, uninterrupted read of approximately 3.2 kilobases in length, by turning the lasers off and on during the run, one could double the size of the read, which consists of around 20 short "strobed" sequence reads that are interspersed with gaps.
- Uninterrupted reads are limited to approximately 3,000 bases due to laser-induced photochemistry that damages the polymerases, he explained. However, when the lights are turned off, the polymerases keep incorporating nucleotides without incurring damage, and the available read length is "essentially put on hold, so you can take whatever read length you have and distribute it" over a longer stretch of DNA
- Researchers could, for example, create the equivalent of a mate-pair read, with two sequences of more than a kilobase at each end and a long stretch of unidentified bases in between. Alternatively, they could cover a long piece of DNA with lots of short stitches of sequence. The length of these stitches and their distance can be varied without a need for different DNA libraries, Turner noted.
- http://www.pacificbiosciences.com/newsletter_2011_05/informatics.php
- http://www.pacbiodevnet.com/ login
- http://www.pacbiodevnet.com/ConversionTools
- SRX032451 SRA dataset; Sequencing for H1 (Haiti, 2010)
- Ecoli Company dataset ; registration needed
- http://www.genomeweb.com/informatics/pacbio-releases-open-source-analysis-suite-ahead-single-molecule-sequencer-launc
Megachile rotundata PacBio sequence
- HDF5 (http://www.hdfgroup.org/HDF5/) This is PacBio's native data format, used to feed their primary and secondary pipelines. There is almost no support for this format outside of PacBio's analysis pipeline. Subsets of this data may be generated, however, as indicated below (e.g., FASTA and FASTQ format). According to the documentation (attached) for PacBio's hybrid assembly protocol (HybridAssembly) you may use HDF5 or FASTA in conjunction with Illumina-based scaffolds (or assembly-rpoduced contigs.)
- High Quality (HQ) FASTA/FASTQ (post-primary analysis) We can derive from the HDF5 file FASTA and FASTQ sequences for reads passing primary analysis in the PacBio pipeline. These reads have essentially been "okayed" by the pipeline as high quality reads to carry forward in secondary analysis--such as attempted alignment to a reference genome, etc. These reads have not had PacBio's proprietary SMRT bell adaptor (45 nt x 2) trimmed... as trimming is part of the downstream PacBio secondary analysis pipeline. These are single pass representations of the sequence reads and therefore contain uncorrected errors, insertions, and deletions incidental to the current state of the PacBio technology. The FASTQ files have PacBio's "Phred-like" quality values corresponding to the nucleotide representation of the sequence. In the instances where a SMRT bell molecule is sequenced multiple times (i.e., multiple passes) each pass is termed a "sub-read". In this representation of the data, sub-reads have not been broken out from the parent sequence.
- High Quality (HQ) FASTA (edited) These FASTA files are similar to the ones described above, but have been filtered and edited. Here, the SMRT bell adaptors have been excised from the parent reads, and entries have been broken into corresponding sub-reads. Only reads with a quality filter of 75% or better have been retained; sub-reads less than 50 nt in length have been removed. These are still single pass representations of the sequence reads and therefore contain uncorrected errors, insertions, and deletions incidental to the current state of the PacBio technology.
Visigen
Polonator
RainDance Technologies
Whole genome profiling (Wgp)
In our view getting to a good quality assembly especially in large genomes is a real challenge and therefore I would like to get your attention to our Whole Genome Profiling technology that generated a lot of interest at the last PAG meeting in San Diego, where we presented the technology in several workshops. WGP has now been used to help the assembly of many of the plant genomes for Melon, Tomato, Potato, Lettuce, Brassica napus, Brassica rapa, Brassica oleraceae, Strawberry, Tobacco, Wheat, and Sunflower. Also at the moment we are working on a number of genomes both for academic labs/consortia as well as for industry. The power of the WGP technology lies in the creation of a sequence-based scaffold of the genome based on the creating of sequence tags in BAC libraries (average 30 tags pet BAC). This provides ordered BAC clones covering the genome on which shot-gun sequence information can be placed. WGP solves the problem of genome assembly that most laboratories are struggling with because it simplifies the bio-informatics effort required to assemble shot-gun sequence information considerably. For instance, shot-gun sequencing of the melon genome (450 Mb) left us with 13,486 contigs, while the WGP map for Melon gave us BAC-based 1085 contigs. By putting these two methods together, we ended up with a very high quality reference amp of only 249 supercontigs. In addition, the BAC clones linked to the physical map allows you to go directly to clones of those regions of the genome you are interested in. I have included three relevant abstracts of PAG presentations in the attachment for your reference. In short: The assembly of the tetraploid tobacco genome (4.5 Gb) was presented at the PAG and worked perfectly well, showing that genome size does not influence the quality of the WGP maps (see abstract below) and in addition showed that the WGP assemblies are very accurate as concluded from the fact that 97% of the WGP contigs can be assigned to one of the ancestral genomes that are part of the tetraploid tobacco genome. Also the progress on the Sunflower genome (3.6 Gb) was presented and my colleague Michiel van Eijk showed the power of the technology to deal with repetitivegenomes as well a new approach on genome sequence assembly that takes as a starting oint a good quality WGP physical map. Taking all this together and the fact that the technical paper on WGP was just accepted forpublication in Genome Research (preprint included in attachment), I think this technology may be of interest for you to improve your genome assembly as well.
Ion Torrent
- SImilar to 454; indel & homopolimer issues
Download
Genbank
- Bioperl scripts:
bp_fetch.pl net::genbank:NC_005810.1 > NC_005810.1 bp_fetch.pl net::genbank:NC_005810 > NC_005810 bp_fetch.pl net::genbank:45439865 > 45439865
- NCBI utilities:
wget "http://eutils.ncbi.nlm.nih.gov/entrez/eutils/epost.fcgi?dbfrom=nucleotide&id=257481614" -O - | grep WebEnv | p 'next unless /<WebEnv>(.+)<\/WebEnv>/; echo "setenv WebEnv $1\n";' wget "http://eutils.ncbi.nlm.nih.gov/entrez/eutils/efetch.fcgi?db=nucleotide&query_key=1&WebEnv=NCID_1_35952872_130.14.18.46_9001_1260305427" -O -
TA
query_tracedb "query text species_code='BACILLUS ANTHRACIS'" > ids.bacillus_anthracis.001
( echo -n "retrieve_tgz fasta quality xml " ; cat ids.bacillus_anthracis.001 | head -40000 | paste -s -d "," ) \
| query_tracedb > bacillus_anthracis.001.1.tgz
( echo -n "retrieve_tgz fasta quality xml " ; cat ids.bacillus_anthracis.001 | head -80000 | tail -40000 | paste -s -d "," ) \
| query_tracedb > bacillus_anthracis.001.2.tgz
...
SRA
Format
FASTA
Utilities:
- Indexing large files
#NCBI utilies: expect a certain id format Ex: ">lcl|12345" formatdb -i prefix.fasta -p F -o T fastacmd -d prefix.fasta -p F -s id1,id2,... fastacmd -d prefix.fasta -p F -i prefix.filter
~/bin/formatdb.pl < prefix.fasta > prefix.fasta.offset ~/bin/fastacmd.pl -s 1,2 -o prefix.fasta.offset < prefix.fasta > subset.fasta
TA
- Sanger:
tarchive2amos -o Ba Ba.seq # TA FTP tarchive2amos -o Ba -tracedir traces/ # TA querytrace_db tarchive2amos -o Ba -assembly assembly/ASSEMBLY.xml -tracedir traces/ # AA
SRA
- Solexa mated:
seq2amos.pl -n solexap -m 100 -s 20 -fs solexa_f.fasta -rs solexa_f.fasta > solexap.afg seq2amos.pl -n solexap -m 100 -s 20 -fs solexa_f.fasta -fq solexa_f.qual -rs solexa_r.fasta -rq solexa_r.qual > solexap.afg seq2amos.pl -n solexap -m 100 -s 20 -fs solexa_f*.fasta -rs solexa_r*.fasta > solexap.afg
- Solexa unmated:
seq2amos.pl -n solexa -fs solexa.fasta > solexa.afg
- 454 mated:
seq2amos.pl -n 454p -m 3000 -s 300 -fs 454.seq > 454p.afg
- 454 unmated:
seq2amos.pl -n 454 -fs 454.seq > 454.afg
Convestion
- Readseq
~/bin//readseq.sh -f Fasta -o prefix.fasta prefix.embl
- Bioperl
bp_sreformat.pl -i prefix.embl -o prefix.fasta -if EMBL -of Fasta
- AMOS:
amos2frg [-i infile] [-o outfile] amos2sq [-i] infile [-o outprefix] => outprefix.seq outprefix.qual
Read Simulators
- Metasim
Read Mapping Software
BFAST
- need to e-mail to author to get the code
BLAT
- BLAT—The BLAST-Like Alignment Tool, Genome Research 2002
- FAQ
- Can align any type of reads
- Can do nt:aa translation
- Command: blat
blat -noHead -t=dna -q=dna -tileSize=10 -stepSize=3 Pa.1con Pa.seq Pa.blat
BLASTALL
- Env vars
echo $BLASTMAT $BLOSUM62 $BLASTDB /fs/sz-user-supported/Linux-x86_64/packages/blast-2.2.18/data/ /fs/sz-user-supported/Linux-x86_64/packages/blast-2.2.18/data/BLOSUM62 /fs/szdata/ncbi/ftp.ncbi.nih.gov/blast/db/
- Example
blastall -p blastn -d refseq_genomic -i acc.keep.fna
BOWTIE
- build index:
bowtie-build prefix.1con prefix => prefix.rev.2.ebwt prefix.rev.1.ebwt prefix.2.ebwt prefix.1.ebwt prefix.3.ebwt prefix.4.ebwt
- run alignments
bowtie prefix qry.fastq -a --best --strata -n 2 -l 28 -e 70
MAQ/BWA
RMAP
- RMAP : designed for Illumina-Solexa
- Command: rmap
rmap -m 3 -w 33 -c Pa.1con Pa.seq -o Pa.rmap
SHRiMP
- Web site
- Commands: rmapper-cs , rmapper-ls, ...
SeqMap
- SeqMap developed at Stanford
- allows up to five mixed substitutions and inserted/deleted nucleotides in the mapping
- allows sequences to contain N’s, and to have unequal lengths
./seqmap Usage: seqmap <number of mismatches> <probe FASTA file name> <transcript FASTA file name> <output file name> [options] Parameters: <number of mismatches> maximum edit distance allowed <probe FASTA file name> probe/tag/read sequences <transcript FASTA file name> reference sequences <output file name> name of the output file ...
SHORE
SOAP
- Web site (China)
- Formatofoutput
- SOAP: short oligonucleotide alignment program, Bioinformatics Jan 2008
- Commands: soap, soap.contig, soap_dealign, soap.huge, soap.short
- can use qualities, do read trimming, use pair ends, RNA alignments
soap -v <max_mismatches> -g <max_gap> -r <repeat_behavior> -p <processors> -d ref.fasta -a qry.fasta -o ref-qry.soap
SOAPaligner/soap2
- ~10 times faster than SOAP
/nfshomes/dpuiu/szdevel/soap2.20release/2bwt-builder ref.fasta # => ref.fasta.index soap2 -D ref.fasta.index -a qry.fasta -o qry.soap -r <repeat_behavior> -l <seed_len> -p <processors> soap2 -D /scratch1/Bombus_impatiens/Data/mito.1con.index -a s_3_1_sequence.seq -o s_3_1_sequence.soap2 -r 2 -l 36 -p 16 # up to 2 mismatches in the first 36bp
SOCS
- Web site
- ABI color space
socs socs.pref more socs.pref Req.fa Seq_F3.csfasta Seq_F3_QV.qual out_prefix 2 1000 2 false true 0
SOLiD
SSAHA
- Web site(Sanger)
- Focused on exact, nearly exact matches
- Does not find all the exact matches???
- Example: Solexa 33bp ~30% of reads are not found
ZOOM
Whole genomes alignments
- BLAT
- BLASTZ, Post-processing long pairwise alignments article. decom program
- LAGAN
- MUMMER
- AVID
Genome Resequencing
- The complete genome of an individual by massively parallel DNA sequencing (J.Watson's genome) Nature April 2008
- J.Watson's genome (supplementary info)
Data sets
Solexa
- Strep suis Solexa at Sanger 2,659,250 36bp single reads, ~49X coverage
/fs/szdata/Solexa/Streptococcus_suis/suisp17/strip3.fna Velvet (0.7.55) assembly: . elem min q1 q2 q3 max mean n50 sum misassembled seq 2659250 36 36 36 36 36 36 36 95733000 . ctg 669 45 69 383 4314 32213 2944 8437 1969901 8 time 153.166u 4.673s 2:20.85 112.0% 0+0k 0+0io 2pf+0w
SOAPdenovo (1.03) assembly: . elem min q1 q2 q3 max mean n50 sum misassembled seq 2659250 36 36 36 36 36 36 36 95733000 ctg 2219 24 25 32 93 24421 907 7209 2014315 1 ctg45 866 45 65 214 3307 24421 2282 7275 1976767 1 time 89.614u 1.599s 1:36.51 94.4% 0+0k 0+0io 0pf+0w
SOAPdenovo vs Velvet: * slightly shorted ctgs * fewer errors (1 vs 8) * shorter run time (~half)
- velvet example: 142,858 35bp reads that should assemble into a ~100Kbp contig
/nfshomes/dpuiu/szdevel/velvet/data/test_reads.fa
- Staphylococcus aureus strain MW2 (edena paper) 35bp, ~47X coverage
- Pseudomonas aeruginosa: 33bp, ~43X coverage
- Pseudomonas syringae: 32bp, ~31X coverage
- 1000 Genomes (June 14th 2008): 47bp
Accession #Runs Instrument Center Study [Individual] SRA000303 41 Solexa 1G Genome Analyzer BI 1000Genomes Project Pilot 2 NA12878 SRA000304 49 Solexa 1G Genome Analyzer BI 1000Genomes Project Pilot 2 NA12891 SRA000305 56 Solexa 1G Genome Analyzer BI 1000Genomes Project Pilot 2 NA12892 SRA000307 1 Solexa 1G Genome Analyzer SC 1000Genomes Project Pilot 1 NA10851 SRA000308 2 Solexa 1G Genome Analyzer SC 1000Genomes Project Pilot 1 NA11993 SRA000309 3 Solexa 1G Genome Analyzer SC 1000Genomes Project Pilot 1 NA11995 SRA000310 1 Solexa 1G Genome Analyzer SC 1000Genomes Project Pilot 1 NA12006 SRA000311 1 Solexa 1G Genome Analyzer SC 1000Genomes Project Pilot 1 NA12044 SRA000312 2 Solexa 1G Genome Analyzer SC 1000Genomes Project Pilot 1 NA12156 SRA000313 1 Solexa 1G Genome Analyzer SC 1000Genomes Project Pilot 1 NA12414 SRA000314 1 Solexa 1G Genome Analyzer SC 1000Genomes Project Pilot 1 NA12776 SRA000315 1 Solexa 1G Genome Analyzer SC 1000Genomes Project Pilot 1 NA12828 SRA000316 12 Solexa 1G Genome Analyzer SC 1000Genomes Project Pilot 2 NA12878 SRA000317 8 Solexa 1G Genome Analyzer SC 1000Genomes Project Pilot 2 NA12891 SRA000318 14 Solexa 1G Genome Analyzer SC 1000Genomes Project Pilot 2 NA12892 SRA000319 1 Solexa 1G Genome Analyzer SC 1000Genomes Project Pilot 1 NA12004
June 14th 2008: Sept 19th 2008
SRA001100 23 Illumina Genome Analyzer BGI 1000Genomes Project Pilot 2 NA19240 ... SRA002029 1 Illumina Genome Analyzer II WUGSC 1000Genomes Project Pilot 2 NA19239
/fs/szdata/Solexa/1000genomes
- Example SRR001113.seq :
7,058,926 47 bp sequences 2,402,398 contain at least 1 '.'
454
- 1000 Genomes
June 14th 2008
Accession #Runs Instrument Center Study [Individual] SRA000302 121 454 GS FLX BCM 1000Genomes Project Pilot 2 NA12878 SRA001032 2 454 GS FLX BCM 1000Genomes Project Pilot 2 NA12878 SRA001036 1 454 GS FLX BCM 1000Genomes Project Pilot 1 NA12812 SRA001094 1 454 GS FLX BCM 1000Genomes Project Pilot 2 NA12878
June 14th 2008: Sept 19th 2008
SRA001037 2 454 GS FLX BCM 1000Genomes Project Pilot 1 NA12812 ... SRA001819 1 454 GS FLX BCM 1000Genomes Project Pilot 2 NA12878
- Cryptosporidium_muris_RN66: SRA001029 (not paired)
- EcoliK12: SRR001355 (paired)
- Porphyromonas_gingivalis_W83: E8YURXS01 (paired)
Refseq
- /fs/szdata/genomes/human_ncbi_build36/ NCBI build36.1 May 2006 (Current build is 36.3 March 2008)
- /fs/szdata/genomes/human_celera_2001_Orig/